
Chapter 9: Detection Methods
9.1: Direct Detection of DNA in Gels
Fluorescent Intercalating Dye Staining
These techniques allow the detection of DNA bands as small as 10 ng in agarose gels.
Staining of DNA in agarose gels can be achieved by including an intercalating dye in the gel or staining the gel after electrophoresis in a dye-containing solution, followed by a washing step known as de-staining to reduce nonspecific staining.
Historically, ethidium bromide-stained gels were photographed with a standard ultraviolet transilluminator at approximately 300 nm using a camera with an orange filter, whereas gels can currently be documented using digital techniques.
When examining a DNA-containing gel under a UV lamp, the eyes and the skin should be protected from UV exposure.
Ethidium bromide is a mutagen and a potential carcinogen. It should be handled according to the Material Safety Data Sheet and safety protection wear should be used while handling the chemical.
Ethidium bromide waste is usually disposed of as hazardous waste and is handled in accordance with laboratory guidelines.
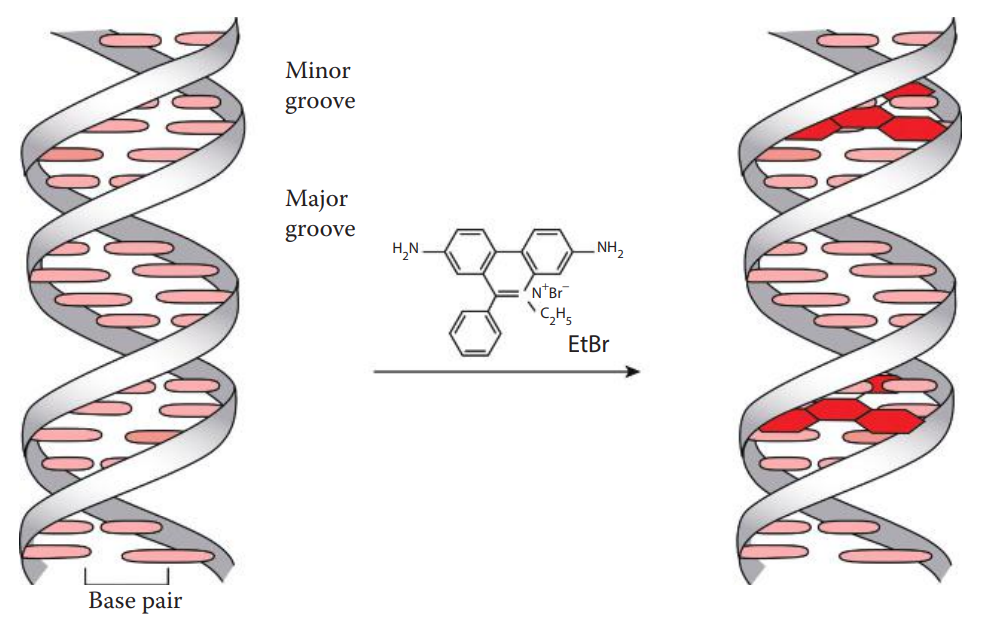
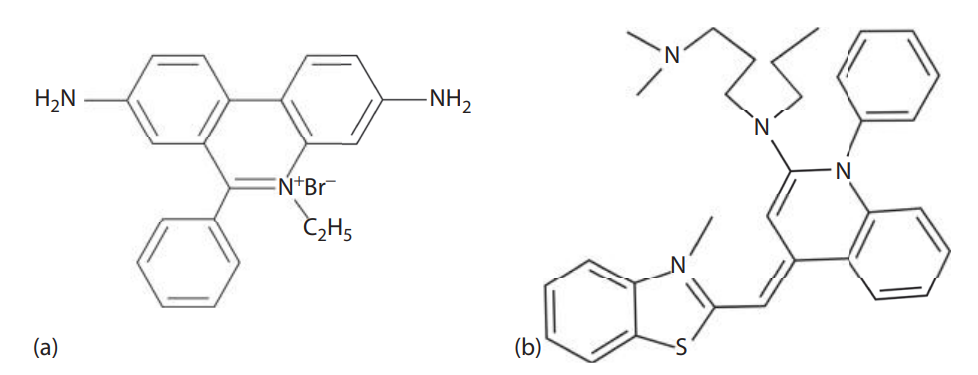
Silver Staining
Silver staining of polyacrylamide gels has been used for the amplified fragment length polymorphism method of variable number tandem repeat profiling.
The sensitivity of silver staining is approximately 100 times higher than that obtained with ethidium bromide, and silver staining is less hazardous than ethidium bromide detection.
Silver staining involves processing a gel followed by exposure to a series of chemicals.
9.2: Detection of DNA Probes in Hybridization-Based Assays
Radioisotope Labeled Probes
Radioisotope probe labeling was used for early versions of VNTR testing and DNA quantitation.
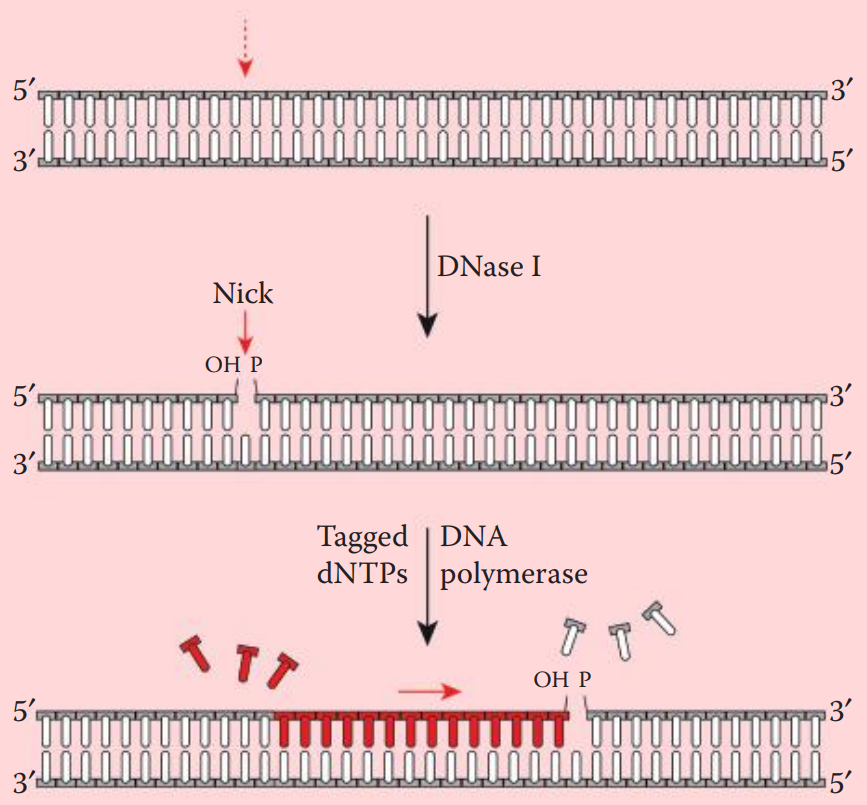
Nick translation incorporates labeled deoxyribonucleotides (dNTPs) into double-stranded DNA.
A DNase is utilized to create nicks along a DNA strand. Tagged nucleotides (one or more than one of the dNTPs) are then incorporated into the DNA strand using a DNA polymerase.
DNA synthesis extends from the 5′ to 3′ end and the original strand is degraded.
DNase I is used to introduce single-strand nicks within the DNA fragment to be labeled.
DNA Polymerase I recognizes the nicks and replaces the preexisting nucleotides with new strands containing labeled dNTPs.
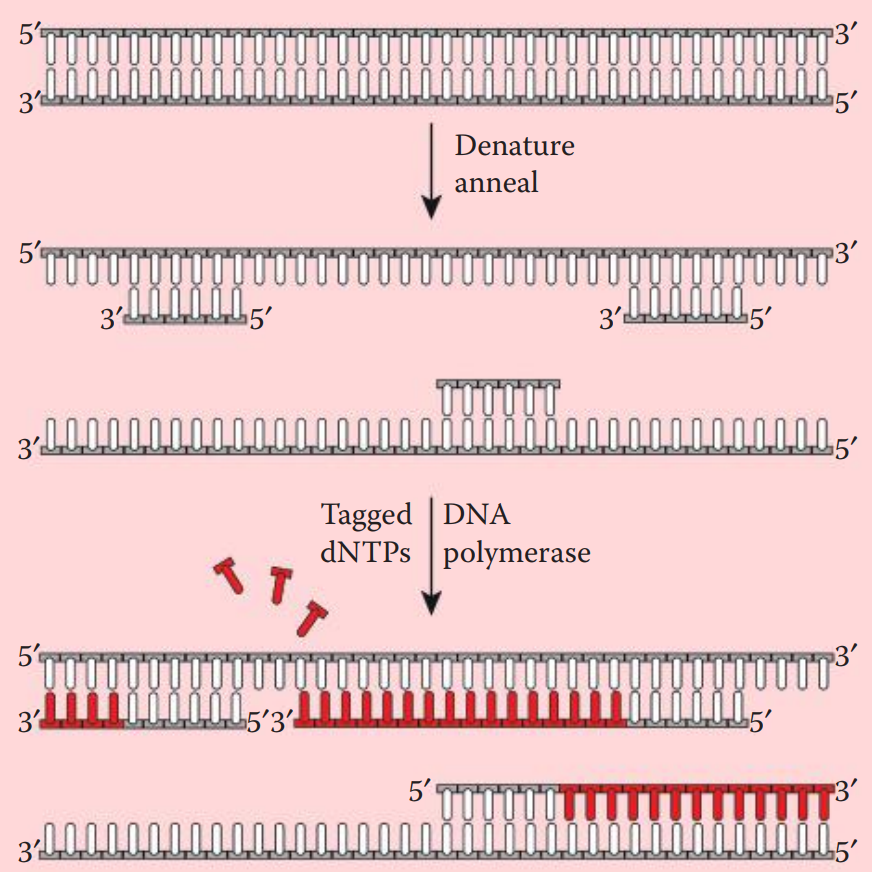
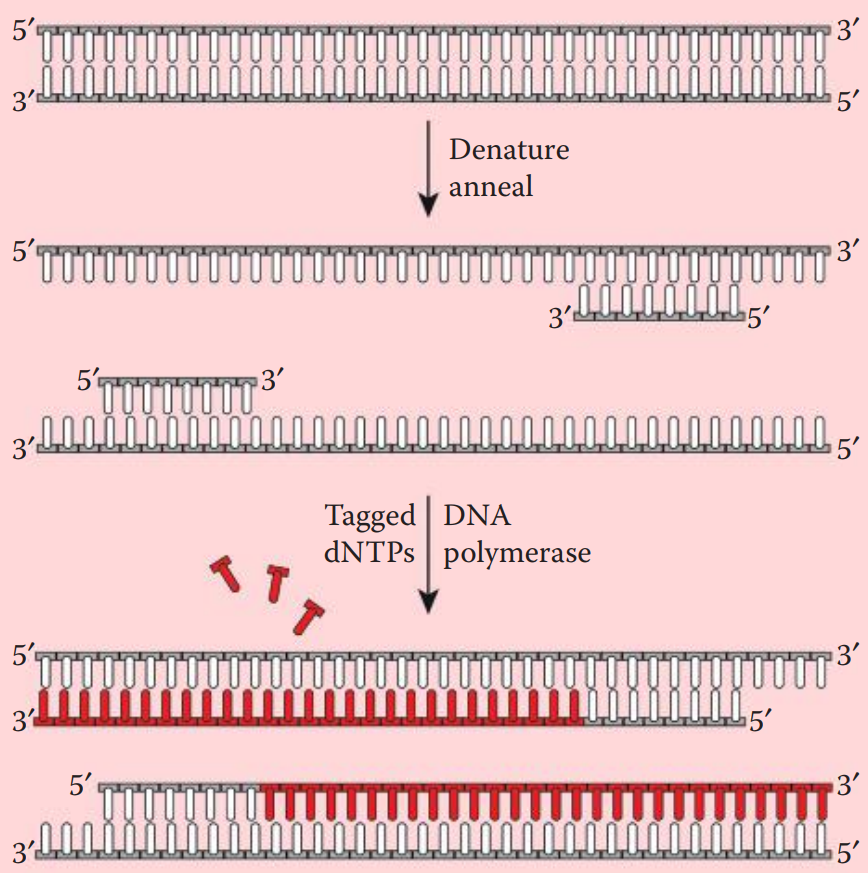
Prior to hybridization, the probe is denatured into single-stranded fragments by boiling for a few minutes followed by rapid cooling on ice.
After the hybridization process, these probes can be visualized by exposing the DNA-containing membrane to an x-ray film.
The energy released from the decay of the radioisotopes is absorbed by the silver halide grains in the film emulsion and forms a latent image.
A chemical development process amplifies the latent image and renders it visible on film.
Enzyme-Conjugated Probe with Chemiluminescence Reporting System
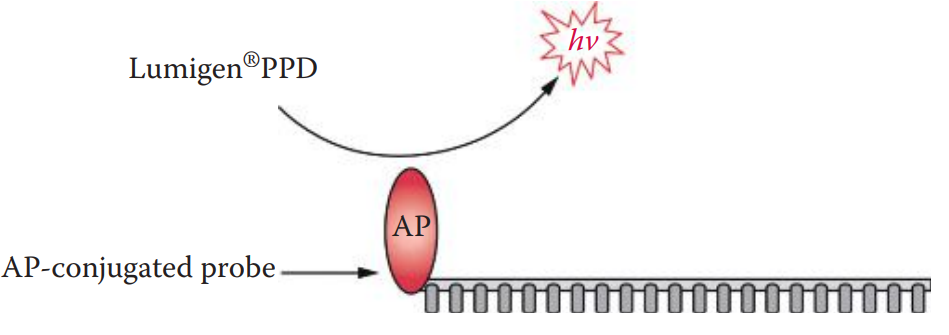
The use of alkaline phosphatase (AP)-conjugated probes with chemiluminescent substrates comprises a highly sensitive non-radioisotopic detection system.
Alkaline phosphatase can cleave the phosphate groups from a variety of substrate molecules.
Its enzymatic activity can be measured using dioxetane-based chemiluminescent substrates such as Lumigen® PPD.
The Lumi-Phos Plus kit of Lumigen Inc. contains this substrate and can serve as a detection system for slot blot assays for DNA quantitation and RFLP assays for VNTR profiling.
AP catalyzes the cleavage of the phosphate ester of Lumigen® PPD, resulting in the release of a photon.
Biotinylation of DNA with Colorimetric Reporting Systems
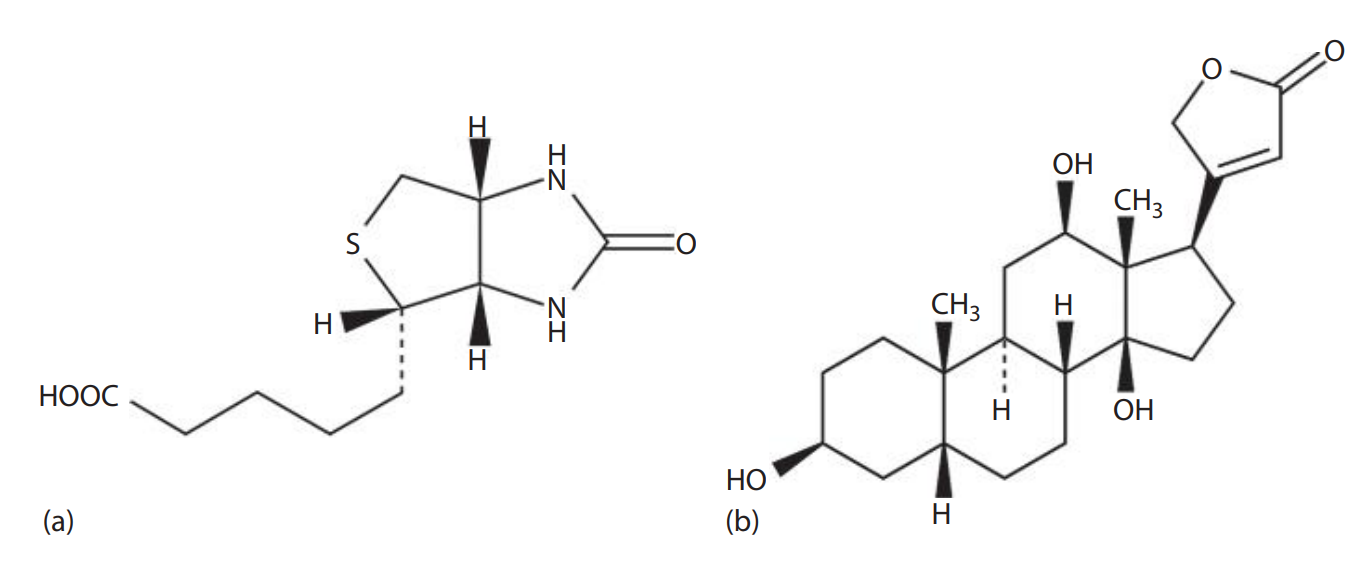
Biotin: Also known as Vitamin H, a water-soluble molecule found in egg yolk.
It can be incorporated into oligonucleotide probes without interfering with the ability of probes to hybridize because of its small size.
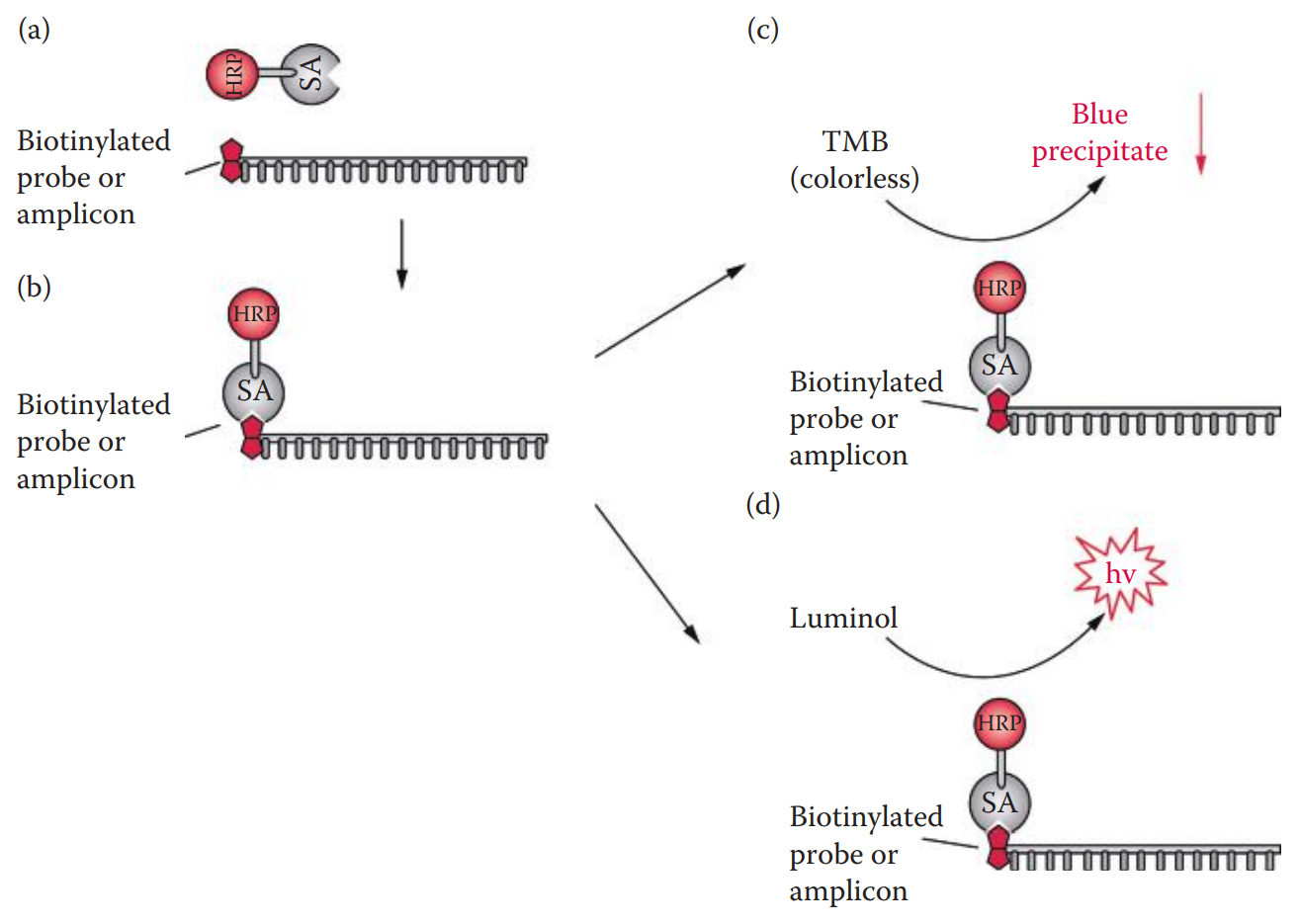
Signals from a biotinylated probe can be detected with an enzyme-conjugated avidin system.
Two steps are required to detect biotin-labeled probes.
First, an avidin conjugate consisting of a reporter enzyme is added.
Then, the reporter enzyme is assayed with substrates.
Enzyme-Conjugated Avidin
Avidin: A glycoprotein found in egg white; it binds to biotin with extremely high affinity.
Avidin detection has a high background due to nonspecific binding.
The nonspecific binding can be reduced by replacing avidin with its streptavidin counterpart from Streptomyces avidin.
Reporter Enzyme Assay
This technique has been used for forensic DNA testing such as slot blot assays for DNA quantitation.
A chemiluminescent substrate such as a luminol-based reagent can also be utilized with HRP.
The peroxidase catalyzes the oxidation of luminol to form a chemiluminescent product.
9.3: Detection Methods for PCR-Based Assays
Fluorescence Labeling
Fluorescent Dyes
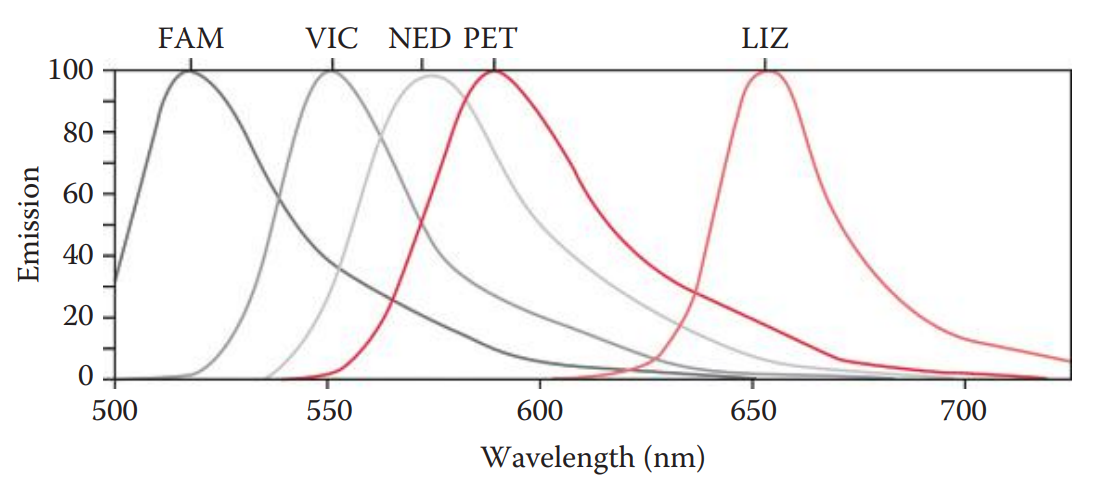
The advantages of fluorescence detection methods include a higher sensitivity and broader dynamic range than comparable colorimetric detection methods.
They have the capacity for simultaneous analysis of complex samples such as multiplex PCR products with different fluorescent labels allowing the distinction of various amplicons.
Labeling Methods
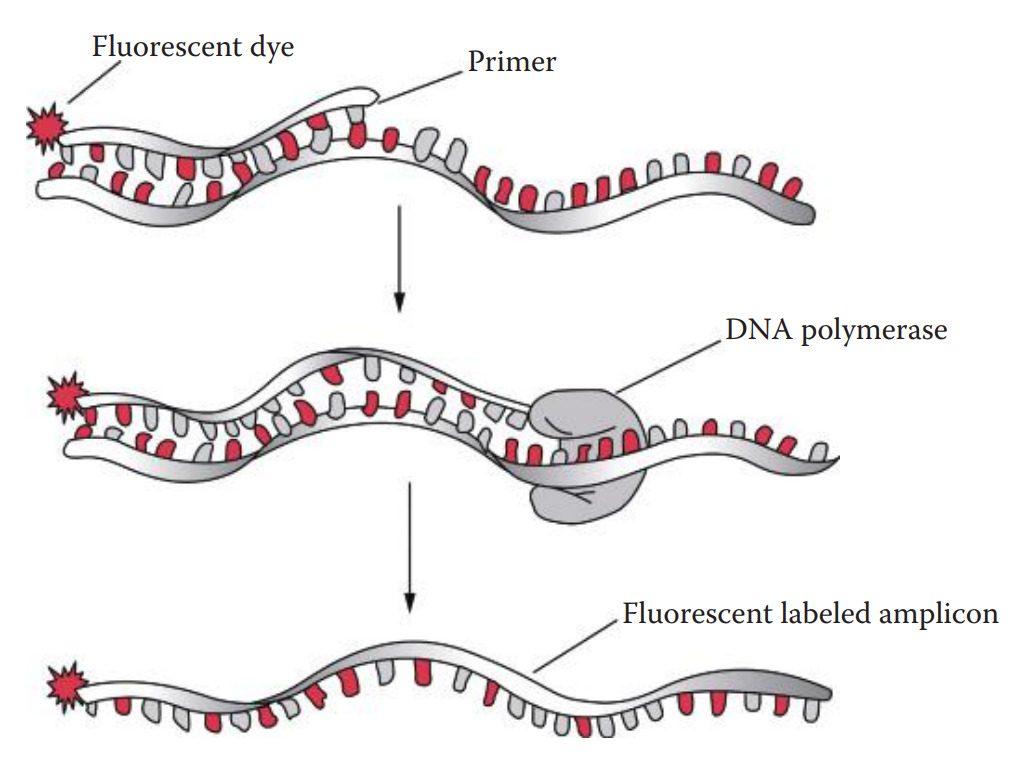
Fluorescent dye labeling can be incorporated into a DNA fragment using a 5′-end fluorescently labeled oligonucleotide primer.
The dye-labeled primer method is usually used for STR profiling in which only one primer from each primer pair is labeled; therefore, only one strand can be detected.
The two-band pattern observed with silver staining does not appear with this method.
Dye-labeled primers allow multiplex PCR amplifications in the same tube.
Fluorescent dye labeling of DNA fragments can be carried out by incorporating fluorescently labeled dideoxynucleotides (ddNTPs) in the PCR product.
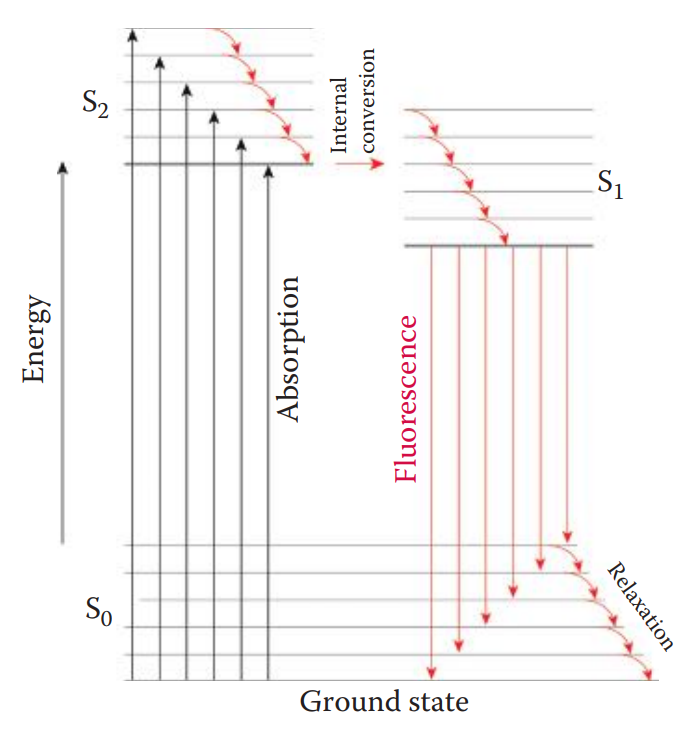
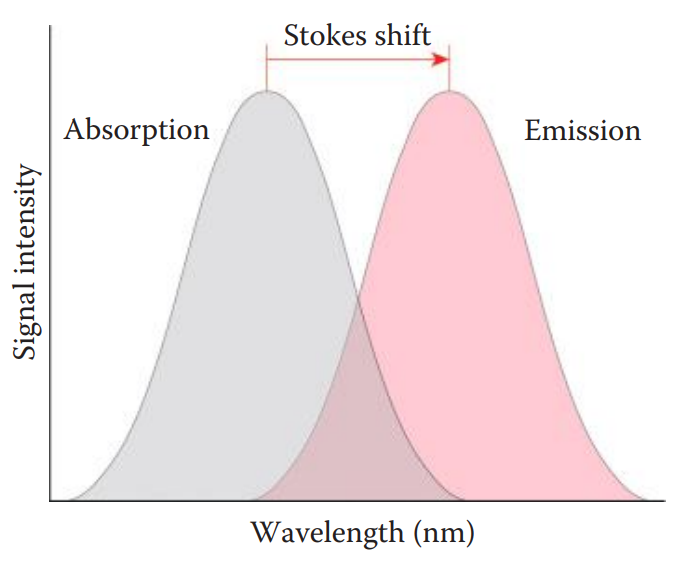
Fluorophore Detection
Fluorophore: A component of a fluorescent dye molecule that causes the molecule to be fluorescent.
Lasers are commonly used as excitation sources because laser light emissions have high intensity and are monochromatic.
The argon ion gas laser is frequently used in applications such as fluorescence-labeled STR and mtDNA sequence analysis because the excitation wavelength of commonly used fluorescent dyes matches the wavelength of the argon laser.
Optical filters are used to filter out undesired light and to allow only one particular wavelength to pass through.
The excitation filter selectively transmits light from an excitation source.
The light is then directed by the dichroic beam splitter to DNA molecules labeled with fluorescent dyes. The light emitted from a fluorophore is also transmitted by the dichroic beam splitter toward the detector.
The emission filter selectively blocks undesired light, thus transmitting a specific wavelength of the emitted fluorescence.
The signal from the fluorophore is collected and converted to an electronic signal expressed in an arbitrary unit such as a relative fluorescence unit (RFU).
Chapter 9: Detection Methods
9.1: Direct Detection of DNA in Gels
Fluorescent Intercalating Dye Staining
These techniques allow the detection of DNA bands as small as 10 ng in agarose gels.
Staining of DNA in agarose gels can be achieved by including an intercalating dye in the gel or staining the gel after electrophoresis in a dye-containing solution, followed by a washing step known as de-staining to reduce nonspecific staining.
Historically, ethidium bromide-stained gels were photographed with a standard ultraviolet transilluminator at approximately 300 nm using a camera with an orange filter, whereas gels can currently be documented using digital techniques.
When examining a DNA-containing gel under a UV lamp, the eyes and the skin should be protected from UV exposure.
Ethidium bromide is a mutagen and a potential carcinogen. It should be handled according to the Material Safety Data Sheet and safety protection wear should be used while handling the chemical.
Ethidium bromide waste is usually disposed of as hazardous waste and is handled in accordance with laboratory guidelines.



Silver Staining
Silver staining of polyacrylamide gels has been used for the amplified fragment length polymorphism method of variable number tandem repeat profiling.
The sensitivity of silver staining is approximately 100 times higher than that obtained with ethidium bromide, and silver staining is less hazardous than ethidium bromide detection.
Silver staining involves processing a gel followed by exposure to a series of chemicals.

9.2: Detection of DNA Probes in Hybridization-Based Assays
Radioisotope Labeled Probes
Radioisotope probe labeling was used for early versions of VNTR testing and DNA quantitation.
Nick translation incorporates labeled deoxyribonucleotides (dNTPs) into double-stranded DNA.
A DNase is utilized to create nicks along a DNA strand. Tagged nucleotides (one or more than one of the dNTPs) are then incorporated into the DNA strand using a DNA polymerase.
DNA synthesis extends from the 5′ to 3′ end and the original strand is degraded.
DNase I is used to introduce single-strand nicks within the DNA fragment to be labeled.
DNA Polymerase I recognizes the nicks and replaces the preexisting nucleotides with new strands containing labeled dNTPs.
Prior to hybridization, the probe is denatured into single-stranded fragments by boiling for a few minutes followed by rapid cooling on ice.
After the hybridization process, these probes can be visualized by exposing the DNA-containing membrane to an x-ray film.
The energy released from the decay of the radioisotopes is absorbed by the silver halide grains in the film emulsion and forms a latent image.
A chemical development process amplifies the latent image and renders it visible on film.



Enzyme-Conjugated Probe with Chemiluminescence Reporting System
The use of alkaline phosphatase (AP)-conjugated probes with chemiluminescent substrates comprises a highly sensitive non-radioisotopic detection system.
Alkaline phosphatase can cleave the phosphate groups from a variety of substrate molecules.
Its enzymatic activity can be measured using dioxetane-based chemiluminescent substrates such as Lumigen® PPD.
The Lumi-Phos Plus kit of Lumigen Inc. contains this substrate and can serve as a detection system for slot blot assays for DNA quantitation and RFLP assays for VNTR profiling.
AP catalyzes the cleavage of the phosphate ester of Lumigen® PPD, resulting in the release of a photon.

Biotinylation of DNA with Colorimetric Reporting Systems
Biotin: Also known as Vitamin H, a water-soluble molecule found in egg yolk.
It can be incorporated into oligonucleotide probes without interfering with the ability of probes to hybridize because of its small size.
Signals from a biotinylated probe can be detected with an enzyme-conjugated avidin system.
Two steps are required to detect biotin-labeled probes.
First, an avidin conjugate consisting of a reporter enzyme is added.
Then, the reporter enzyme is assayed with substrates.

Enzyme-Conjugated Avidin
Avidin: A glycoprotein found in egg white; it binds to biotin with extremely high affinity.
Avidin detection has a high background due to nonspecific binding.
The nonspecific binding can be reduced by replacing avidin with its streptavidin counterpart from Streptomyces avidin.
Reporter Enzyme Assay
This technique has been used for forensic DNA testing such as slot blot assays for DNA quantitation.
A chemiluminescent substrate such as a luminol-based reagent can also be utilized with HRP.
The peroxidase catalyzes the oxidation of luminol to form a chemiluminescent product.

9.3: Detection Methods for PCR-Based Assays
Fluorescence Labeling
Fluorescent Dyes
The advantages of fluorescence detection methods include a higher sensitivity and broader dynamic range than comparable colorimetric detection methods.
They have the capacity for simultaneous analysis of complex samples such as multiplex PCR products with different fluorescent labels allowing the distinction of various amplicons.

Labeling Methods
Fluorescent dye labeling can be incorporated into a DNA fragment using a 5′-end fluorescently labeled oligonucleotide primer.
The dye-labeled primer method is usually used for STR profiling in which only one primer from each primer pair is labeled; therefore, only one strand can be detected.
The two-band pattern observed with silver staining does not appear with this method.
Dye-labeled primers allow multiplex PCR amplifications in the same tube.
Fluorescent dye labeling of DNA fragments can be carried out by incorporating fluorescently labeled dideoxynucleotides (ddNTPs) in the PCR product.

Fluorophore Detection
Fluorophore: A component of a fluorescent dye molecule that causes the molecule to be fluorescent.
Lasers are commonly used as excitation sources because laser light emissions have high intensity and are monochromatic.
The argon ion gas laser is frequently used in applications such as fluorescence-labeled STR and mtDNA sequence analysis because the excitation wavelength of commonly used fluorescent dyes matches the wavelength of the argon laser.
Optical filters are used to filter out undesired light and to allow only one particular wavelength to pass through.
The excitation filter selectively transmits light from an excitation source.
The light is then directed by the dichroic beam splitter to DNA molecules labeled with fluorescent dyes. The light emitted from a fluorophore is also transmitted by the dichroic beam splitter toward the detector.
The emission filter selectively blocks undesired light, thus transmitting a specific wavelength of the emitted fluorescence.
The signal from the fluorophore is collected and converted to an electronic signal expressed in an arbitrary unit such as a relative fluorescence unit (RFU).


 Knowt
Knowt
