
Chapter 7: Amplification by Polymerase Chain Reaction
7.1: Denaturation and Renaturation of DNA
The DNA double helix is stabilized by chemical interactions.
An adenine always pairs with a thymine (two hydrogen bonds), and a cytosine pairs with guanine (three hydrogen bonds).
Double-stranded DNA is maintained by hydrogen bonding between the bases of complementary pairs.
Denaturation: Occurs when the hydrogen bonds of DNA are disrupted, and the strands are separated.
Melting Curve: Can be obtained from measuring DNA denaturation by slowly heat- ing a solution of DNA.
Melting Temperature: The temperature at which 50% of DNA strands are denatured.
Renaturation: The single strands in a solution of denatured DNA can reanneal into double-stranded DNA.
7.2: Basic Principles of Polymerase Chain Reaction
The polymerase chain reaction (PCR) allows the exponential amplification of specific sequences of DNA to yield sufficient amplified products, also known as amplicons, for various downstream applications.
PCR forms the basis of many forensic DNA assays such as DNA quantitation, STR profiling and mtDNA sequencing.
Real-Time PCR: This process allows the monitoring of amplicon production at each cycle of the PCR process.
An S-shaped amplification curve is obtained and divided into an exponential phase, a linear phase, and a plateau.
7.3: Essential PCR Components
Thermostable DNA Polymerases
Taq polymerase is the most commonly used enzyme for routine PCR applications.
AmpliTaq Gold DNA polymerase (Applied Biosystems) is the most common DNA polymerase for forensic applications.
AmpliTaq Gold DNA polymerase: A modified enzyme, remains in an inactive form until activated with a pH below 7 prior to the PCR cycling in which the inhibitory motif is inactivated.
The pH of the buffer system used in the PCR reaction is temperature sensitive; increasing the temperature decreases the pH of the solution.
PCR Primers
PCR primers are the oligonucleotides that are complementary to the sequences that flank the target region of the template. A pair of primers is required.
Properly designed primers are critical to the success of a PCR reaction. Computer software such as Primer3 is available to assist and optimize the design of primers.
A primer must be specific to the target sequence; otherwise, nonspecific products that might interfere with the proper interpretation of a DNA profile might be produced.
A primer should not contain self-complementary sequences that may form hairpin structures interfering with the annealing of primers and the template.
These annealed primers may then be amplified during PCR, creating products known as primer dimers, which compete with the target DNA template for PCR components.
Multiplex PCR is the simultaneous amplification of more than one region of a DNA template in a single reaction to achieve high-throughput analysis.
Multiplex PCR consists of multiple primer sets in a single reaction to produce amplicons of multiple target DNA regions.
Other Components
Essential components include template DNA with target sequences in either linear form (nuclear genomic DNA) or circular form (mitochondrial DNA).
Both single- and double-stranded DNA can be used as a template for PCR.
Deoxynucleoside triphosphates (dNTPs): The substrates for DNA synthesis.
Divalent cations are required for the enzymatic activity of DNA polymerases.
Monovalent cations are usually recommended, and a buffer is often utilized to maintain pH between 8.3 and 8.8 at room temperature.
Controls should be used to monitor the effectiveness of PCR amplification.
A positive control shows that PCR components such as reagents and PCR cycle parameters are working properly during a PCR.
7.4: Cycle Parameters
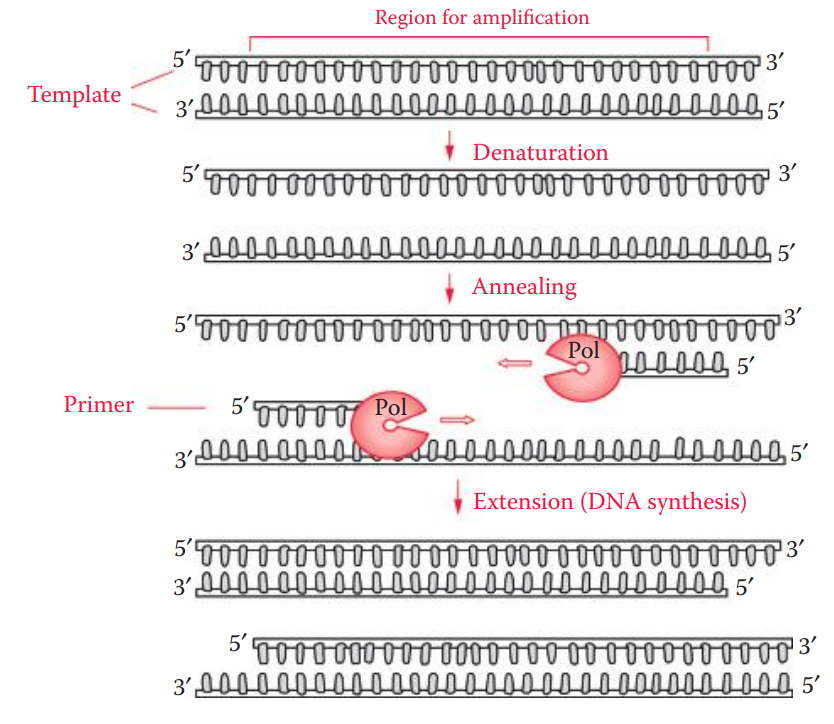
PCR cycle consists of three elements: denaturation, annealing, and extension.
At the beginning of each cycle, the two complementary DNA template strands are separated at high temperatures (94°C–95°C) in a process called denaturation.
The temperature is then decreased to allow annealing between the oligonucleotide primers and the template. The temperature for annealing is usually 3°C–5°C.
If the annealing temperature is too high, a very low quantity of amplicon is yielded because of the failure of annealing between the primer and the template.
If the annealing temperature is too low, nonspecific amplification can occur.
Optimal temperature for DNA polymerase is reached, thus allowing for DNA replication (extension).
Thermal Cycler: Instrument that carries out PCR amplification.
7.5: Factors Affecting PCR
Template Quality
It is important to prevent the degradation of the DNA during the collection and processing of evidence. Degradation causes DNA to break into smaller fragments.
If the damage occurs at a region to be amplified, the result can be a failure in PCR amplification.
Stochastic Effect: The two alleles in a heterozygous individual are unequally detected at a low level of starting DNA template.
Inhibitors
Inhibitors can interact with the DNA template or polymerase, causing PCR amplification failure.
The presence of PCR inhibitors can be detected using an internal positive control.
A number of PCR inhibitors commonly encountered in evidence samples include heme molecules from the blood, indigo dyes from fabrics, and melanin from hair samples.
If PCR inhibitors are not eliminated during the extraction process, additional procedures such as the use of centrifugal filtration devices can be used.
Centrifugal filtration devices can separate molecules by size.
Contamination
To prevent contamination, pre-PCR and post-PCR samples should be processed in separate areas or at different times.
Reagents, supplies, and equipment used for pre-PCR and post-PCR steps should be separated as well.
Protective gear should include laboratory coats and disposable gloves. Facial masks and hair caps may be used if necessary.
Aerosol-resistant pipet tips and DNA-free solutions and test tubes should also be used.
Extraction reagent blanks, which contain all extraction reagents but no sample, monitor contamination from extraction to PCR.
Amplification-negative controls, which contain all PCR reagents and no DNA template, monitor contamination of the amplification.
7.6: Reverse Transcriptase PCR for RNA-Based Assays
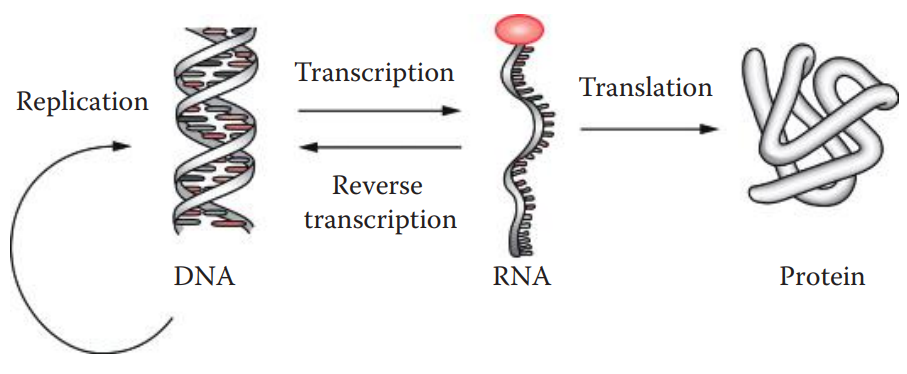
Central dogma: The pathway for the flow of genetic information.
The parental DNA serves as the template for DNA replication.
With RNA synthesis or transcription, the process is carried out using the DNA as a template.
RNA chains can be used as templates for the synthesis of a DNA strand of complementary sequence, in which the end product is referred to as complementary DNA (cDNA).
Protein synthesis, also known as translation, is directed by an RNA template.
Reverse Transcription: The flow of genetic information from RNA to DNA.
Reverse transcription is carried out by a reverse transcriptase that forms the basis of reverse transcriptase PCR.
It is highly sensitive and can be used to detect very small quantities of mRNA. It can be utilized to measure levels of gene expression even when the RNA of interest is expressed at very low levels.
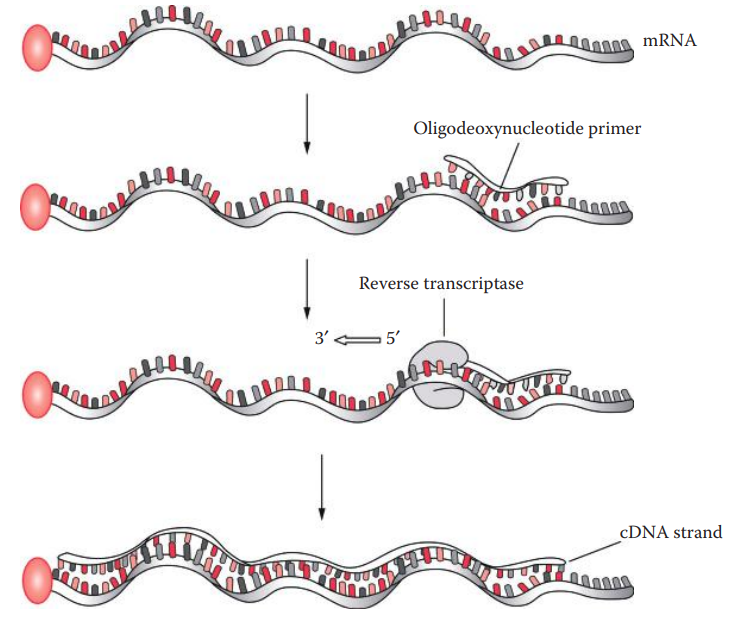
Reverse Transcription
The catalytic function of cDNA synthesis requires a primer that anneals to a complementary mRNA template.
The primer can be either RNA or DNA; however, DNA primers are more efficient than RNA primers.
During the elongation of the primer, reverse transcriptase incorporates the corresponding deoxyribonucleotide triphosphate according to the rules of base pairing with the RNA template.
The RNA template is then degraded by an intrinsic RNase H activity of reverse transcriptase during the reverse transcription reaction.
Retroviral RNase H: A domain of the viral reverse transcriptase enzyme. It is a nonspecific endonuclease that cleaves RNA.
The most common reverse transcriptases used for cDNA synthesis are encoded by the pol gene from the avian myeloblastosis virus (AMV) and the Moloney strain of the murine leukemia virus (MMLV).
Oligodeoxynucleotide Priming
Oligodeoxynucleotide primers are an essential component for a reverse transcriptase reaction.
Gene-specific primers are designed to hybridize to a particular mRNA sequence for the conversion of a specific gene sequence into cDNA.
Universal primers can hybridize to any mRNA sequence in a sample to convert all mRNAs to cDNA.
Oligo (dT) primer can hybridize to the 3′ termini poly (A) tails of eukaryotic mRNAs.
Random hexamer primers are nonspecific primers that can hybridize, at multiple sites, to any RNA sequence including non-mRNA templates such as ribosomal RNA.
Reverse Transcriptase PCR
During an RT-PCR process, a single-stranded cDNA is synthesized from a template mRNA using reverse transcription. The cDNA is then amplified by PCR with a pair of oligonucleotide primers corresponding to a specific sequence in the cDNA,
Two strategies of RT-PCR exist: a one-step and a two-step RT-PCR.
One-step RT-PCR combines the reverse transcription reaction and PCR in a single tube.
During a two-step PCR, the reverse transcription reaction and PCR are carried out in separate tubes.
The cDNA is transferred to a separate tube for PCR amplification.
Two types of PCR methods can be utilized for analyzing amplified products.
The end-point PCR method measures the amount of amplified product synthesized during PCR at the end of the PCR amplification.
With the real-time PCR method, the amplified product is quantified during the exponential phase of PCR.
The hot-start PCR strategy is typically used to increase sensitivity, specificity, and yield.
Chapter 7: Amplification by Polymerase Chain Reaction
7.1: Denaturation and Renaturation of DNA
The DNA double helix is stabilized by chemical interactions.
An adenine always pairs with a thymine (two hydrogen bonds), and a cytosine pairs with guanine (three hydrogen bonds).
Double-stranded DNA is maintained by hydrogen bonding between the bases of complementary pairs.
Denaturation: Occurs when the hydrogen bonds of DNA are disrupted, and the strands are separated.
Melting Curve: Can be obtained from measuring DNA denaturation by slowly heat- ing a solution of DNA.
Melting Temperature: The temperature at which 50% of DNA strands are denatured.
Renaturation: The single strands in a solution of denatured DNA can reanneal into double-stranded DNA.
7.2: Basic Principles of Polymerase Chain Reaction
The polymerase chain reaction (PCR) allows the exponential amplification of specific sequences of DNA to yield sufficient amplified products, also known as amplicons, for various downstream applications.
PCR forms the basis of many forensic DNA assays such as DNA quantitation, STR profiling and mtDNA sequencing.
Real-Time PCR: This process allows the monitoring of amplicon production at each cycle of the PCR process.
An S-shaped amplification curve is obtained and divided into an exponential phase, a linear phase, and a plateau.
7.3: Essential PCR Components
Thermostable DNA Polymerases
Taq polymerase is the most commonly used enzyme for routine PCR applications.
AmpliTaq Gold DNA polymerase (Applied Biosystems) is the most common DNA polymerase for forensic applications.
AmpliTaq Gold DNA polymerase: A modified enzyme, remains in an inactive form until activated with a pH below 7 prior to the PCR cycling in which the inhibitory motif is inactivated.
The pH of the buffer system used in the PCR reaction is temperature sensitive; increasing the temperature decreases the pH of the solution.
PCR Primers
PCR primers are the oligonucleotides that are complementary to the sequences that flank the target region of the template. A pair of primers is required.
Properly designed primers are critical to the success of a PCR reaction. Computer software such as Primer3 is available to assist and optimize the design of primers.
A primer must be specific to the target sequence; otherwise, nonspecific products that might interfere with the proper interpretation of a DNA profile might be produced.
A primer should not contain self-complementary sequences that may form hairpin structures interfering with the annealing of primers and the template.
These annealed primers may then be amplified during PCR, creating products known as primer dimers, which compete with the target DNA template for PCR components.
Multiplex PCR is the simultaneous amplification of more than one region of a DNA template in a single reaction to achieve high-throughput analysis.
Multiplex PCR consists of multiple primer sets in a single reaction to produce amplicons of multiple target DNA regions.
Other Components
Essential components include template DNA with target sequences in either linear form (nuclear genomic DNA) or circular form (mitochondrial DNA).
Both single- and double-stranded DNA can be used as a template for PCR.
Deoxynucleoside triphosphates (dNTPs): The substrates for DNA synthesis.
Divalent cations are required for the enzymatic activity of DNA polymerases.
Monovalent cations are usually recommended, and a buffer is often utilized to maintain pH between 8.3 and 8.8 at room temperature.
Controls should be used to monitor the effectiveness of PCR amplification.
A positive control shows that PCR components such as reagents and PCR cycle parameters are working properly during a PCR.
7.4: Cycle Parameters
PCR cycle consists of three elements: denaturation, annealing, and extension.
At the beginning of each cycle, the two complementary DNA template strands are separated at high temperatures (94°C–95°C) in a process called denaturation.
The temperature is then decreased to allow annealing between the oligonucleotide primers and the template. The temperature for annealing is usually 3°C–5°C.
If the annealing temperature is too high, a very low quantity of amplicon is yielded because of the failure of annealing between the primer and the template.
If the annealing temperature is too low, nonspecific amplification can occur.
Optimal temperature for DNA polymerase is reached, thus allowing for DNA replication (extension).
Thermal Cycler: Instrument that carries out PCR amplification.

7.5: Factors Affecting PCR
Template Quality
It is important to prevent the degradation of the DNA during the collection and processing of evidence. Degradation causes DNA to break into smaller fragments.
If the damage occurs at a region to be amplified, the result can be a failure in PCR amplification.
Stochastic Effect: The two alleles in a heterozygous individual are unequally detected at a low level of starting DNA template.
Inhibitors
Inhibitors can interact with the DNA template or polymerase, causing PCR amplification failure.
The presence of PCR inhibitors can be detected using an internal positive control.
A number of PCR inhibitors commonly encountered in evidence samples include heme molecules from the blood, indigo dyes from fabrics, and melanin from hair samples.
If PCR inhibitors are not eliminated during the extraction process, additional procedures such as the use of centrifugal filtration devices can be used.
Centrifugal filtration devices can separate molecules by size.
Contamination
To prevent contamination, pre-PCR and post-PCR samples should be processed in separate areas or at different times.
Reagents, supplies, and equipment used for pre-PCR and post-PCR steps should be separated as well.
Protective gear should include laboratory coats and disposable gloves. Facial masks and hair caps may be used if necessary.
Aerosol-resistant pipet tips and DNA-free solutions and test tubes should also be used.
Extraction reagent blanks, which contain all extraction reagents but no sample, monitor contamination from extraction to PCR.
Amplification-negative controls, which contain all PCR reagents and no DNA template, monitor contamination of the amplification.
7.6: Reverse Transcriptase PCR for RNA-Based Assays
Central dogma: The pathway for the flow of genetic information.
The parental DNA serves as the template for DNA replication.
With RNA synthesis or transcription, the process is carried out using the DNA as a template.
RNA chains can be used as templates for the synthesis of a DNA strand of complementary sequence, in which the end product is referred to as complementary DNA (cDNA).
Protein synthesis, also known as translation, is directed by an RNA template.
Reverse Transcription: The flow of genetic information from RNA to DNA.
Reverse transcription is carried out by a reverse transcriptase that forms the basis of reverse transcriptase PCR.
It is highly sensitive and can be used to detect very small quantities of mRNA. It can be utilized to measure levels of gene expression even when the RNA of interest is expressed at very low levels.

Reverse Transcription
The catalytic function of cDNA synthesis requires a primer that anneals to a complementary mRNA template.
The primer can be either RNA or DNA; however, DNA primers are more efficient than RNA primers.
During the elongation of the primer, reverse transcriptase incorporates the corresponding deoxyribonucleotide triphosphate according to the rules of base pairing with the RNA template.
The RNA template is then degraded by an intrinsic RNase H activity of reverse transcriptase during the reverse transcription reaction.
Retroviral RNase H: A domain of the viral reverse transcriptase enzyme. It is a nonspecific endonuclease that cleaves RNA.
The most common reverse transcriptases used for cDNA synthesis are encoded by the pol gene from the avian myeloblastosis virus (AMV) and the Moloney strain of the murine leukemia virus (MMLV).
Oligodeoxynucleotide Priming
Oligodeoxynucleotide primers are an essential component for a reverse transcriptase reaction.
Gene-specific primers are designed to hybridize to a particular mRNA sequence for the conversion of a specific gene sequence into cDNA.
Universal primers can hybridize to any mRNA sequence in a sample to convert all mRNAs to cDNA.
Oligo (dT) primer can hybridize to the 3′ termini poly (A) tails of eukaryotic mRNAs.
Random hexamer primers are nonspecific primers that can hybridize, at multiple sites, to any RNA sequence including non-mRNA templates such as ribosomal RNA.
Reverse Transcriptase PCR
During an RT-PCR process, a single-stranded cDNA is synthesized from a template mRNA using reverse transcription. The cDNA is then amplified by PCR with a pair of oligonucleotide primers corresponding to a specific sequence in the cDNA,
Two strategies of RT-PCR exist: a one-step and a two-step RT-PCR.
One-step RT-PCR combines the reverse transcription reaction and PCR in a single tube.
During a two-step PCR, the reverse transcription reaction and PCR are carried out in separate tubes.
The cDNA is transferred to a separate tube for PCR amplification.
Two types of PCR methods can be utilized for analyzing amplified products.
The end-point PCR method measures the amount of amplified product synthesized during PCR at the end of the PCR amplification.
With the real-time PCR method, the amplified product is quantified during the exponential phase of PCR.
The hot-start PCR strategy is typically used to increase sensitivity, specificity, and yield.

 Knowt
Knowt
