
Chapter 6: DNA Quantitation
6.1: Slot Blot Assay
Historically, the slot blot assay was used to detect human genomic DNA in a sample.
Prior to the quantitation, an alkaline solution is added to the genomic DNA sample, which denatures DNA.
Generating single-stranded DNA is necessary for DNA to be cross-linked onto a nitrocellulose membrane.
Three detecting schemes of the slot blot assay have been developed:
D17Z1 probe: Labeled with radioisotopes that could be visualized by exposing the slot blot membrane to x-ray film.
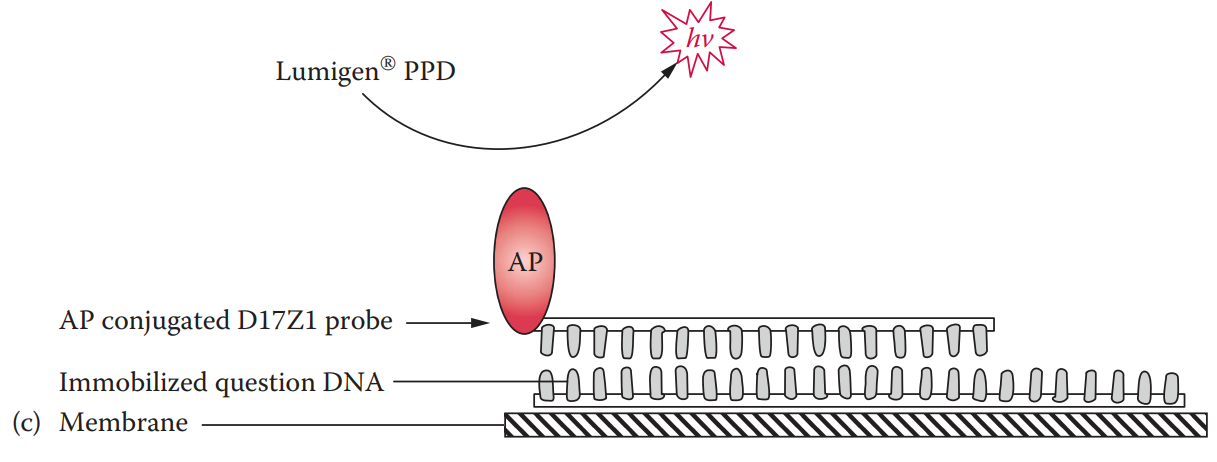
Alkaline phosphatase-labeled probe: It can be coupled with chemiluminescent detection.
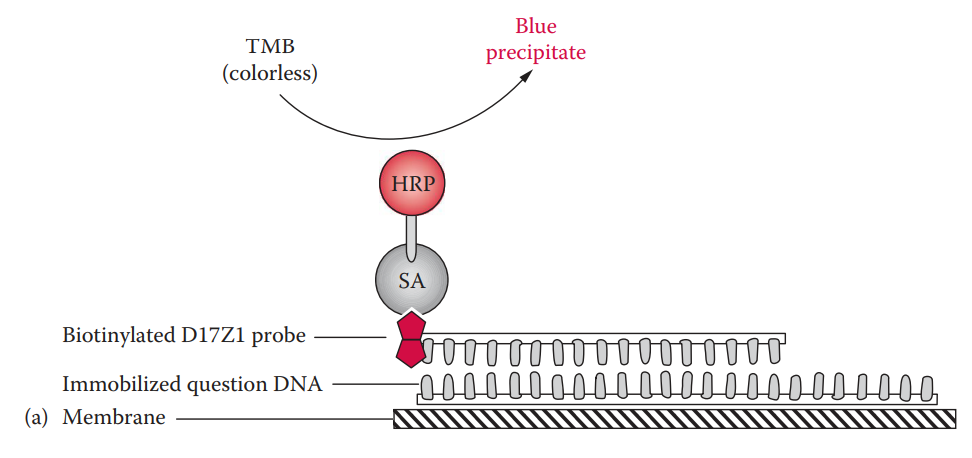
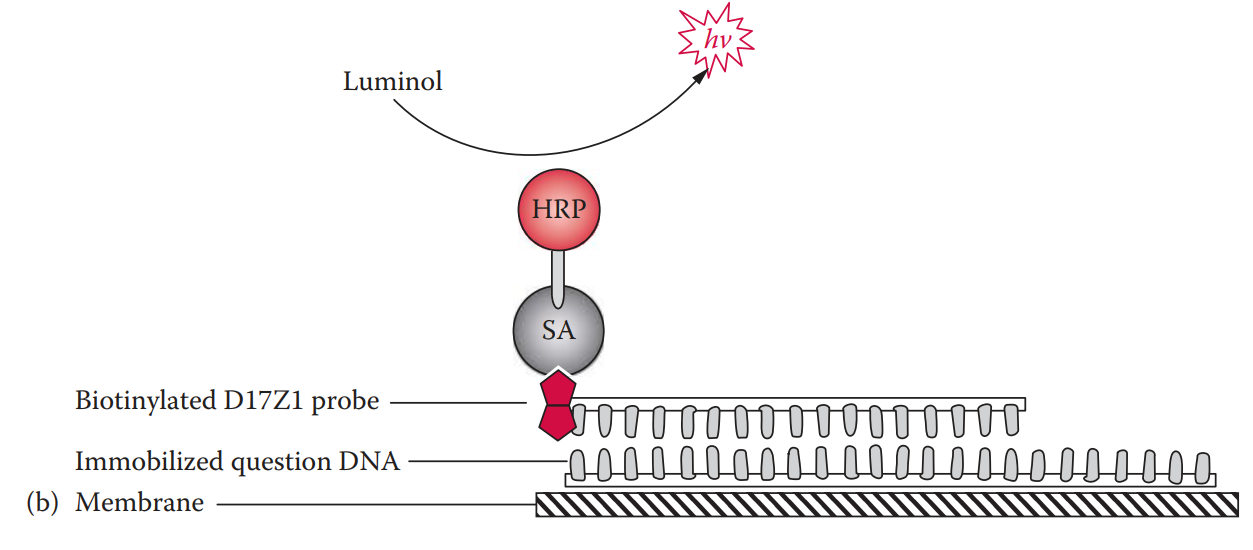
Biotinylated probe: It can be coupled with either colorimetric or chemiluminescent detection.
In colorimetric detection, the biotin moiety of the probe is bound to streptavidin.
The streptavidin is conjugated with horseradish peroxidase, which catalyzes the oxidation reaction of tetramethylbenzidine (TMB), a substrate, forming a blue precipitate.
With the chemiluminescent detection method, the horseradish peroxidase catalyzes the oxidation reaction of a substrate such as luminol, emitting photons that can be detected by exposure to x-ray film.
The sensitivity of chemiluminescent detection is slightly higher than that of colorimetric detection.
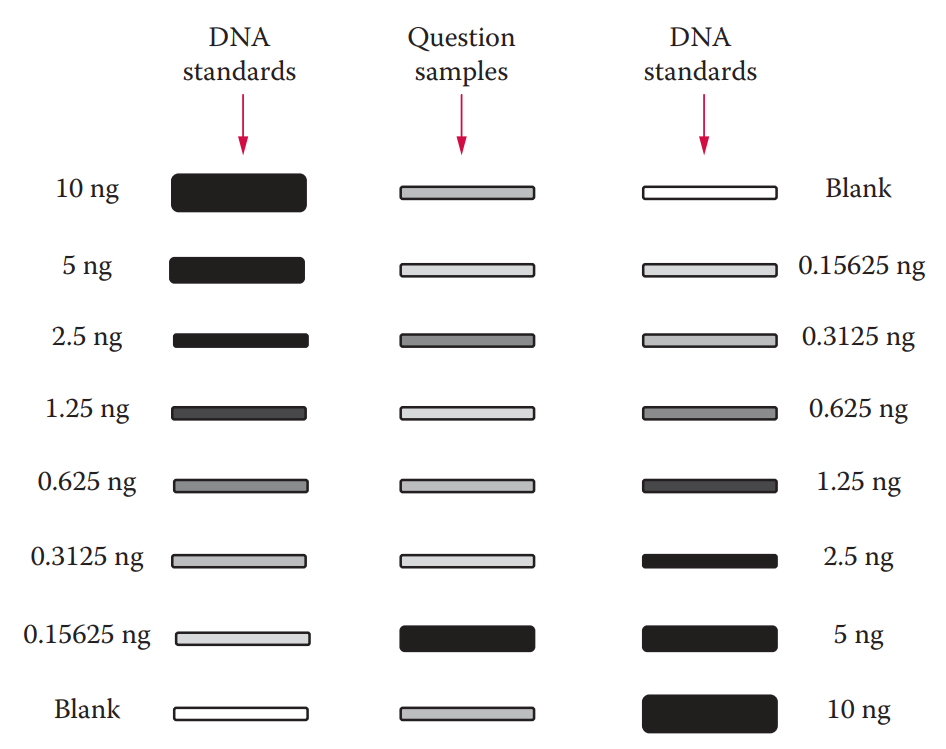
The detected signal intensity is proportional to the concentration of the DNA sample in question. Quantitative measurements can be made by comparing an unknown sample to a set of standards with known DNA concentrations.
6.2: Fluorescent Intercalating Dye Assay
Intercalating dyes, usually planar molecules, can slide themselves in between base pairs of DNA without breaking the DNA double helix.
The Quant-iT PicoGreen dsDNA reagent (Invitrogen) is a fluorescent intercalating dye that stains double-stranded DNA (dsDNA) for quantitation in a sample. The detection limit of this method is approximately 250 pg.
Fluorescent intercalating dye assay can be utilized for the quantitation of known reference samples.
DNA samples are simply added to a solution containing the fluorescent intercalating dye. The fluorescence, proportional to the quantities of DNA is measured using a standard spectrofluorometer with excitation and emission wavelengths of the light source.
A standard curve is first created using samples containing known amounts of DNA. The assay is then performed for the unknown samples and the quantities of DNA in the samples are determined by comparing the results to the standard curve.
6.3: Quantitative PCR Assay
End-point PCR methods measure the quantity of amplified product at the end of PCR.
Real-time PCR methods can quantify the amplified DNA during the exponential phase of PCR.
Real-Time Quantitative PCR
Real-time quantitative PCR (qPCR) was developed in the early 1990s, and it analyzes the amplification of a target sequence at each cycle of PCR.
Fluorescent reporter: Used to monitor the accumulation of amplified products during PCR.
The fluorescence signals of the reporter molecule increase as amplified products accumulate with each cycle of PCR.
qPCR is commonly used because of the following advantages:
Better objectivity than the QuantiBlot method.
Increased sensitivity with a large dynamic range (30 pg–100 ng).
More accurate measurements of small quantities of DNA in samples.
Fewer laboratory manipulations; are amenable to automation.
Ability to detect PCR inhibitors.
The technique is amenable to multiplexing to detect more than one type of DNA target sequence in a single reaction.
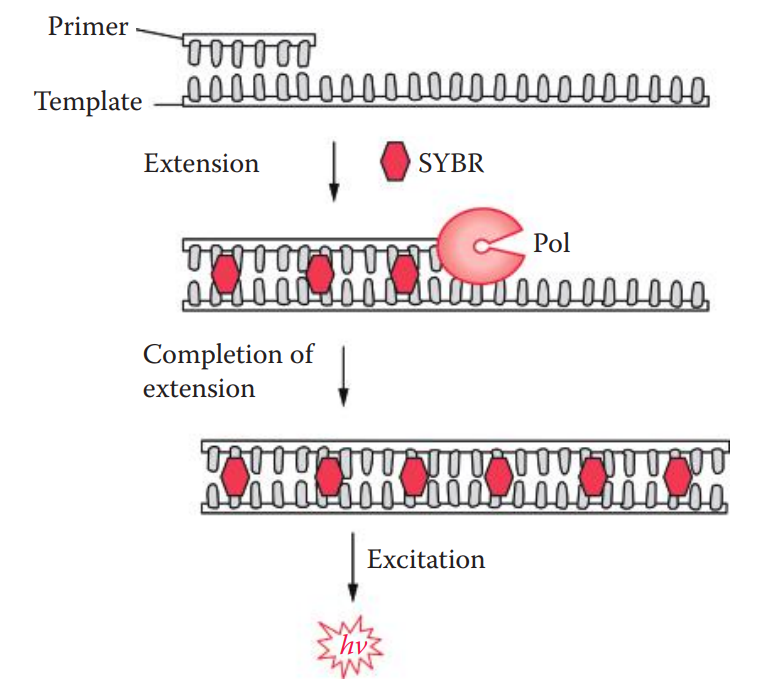
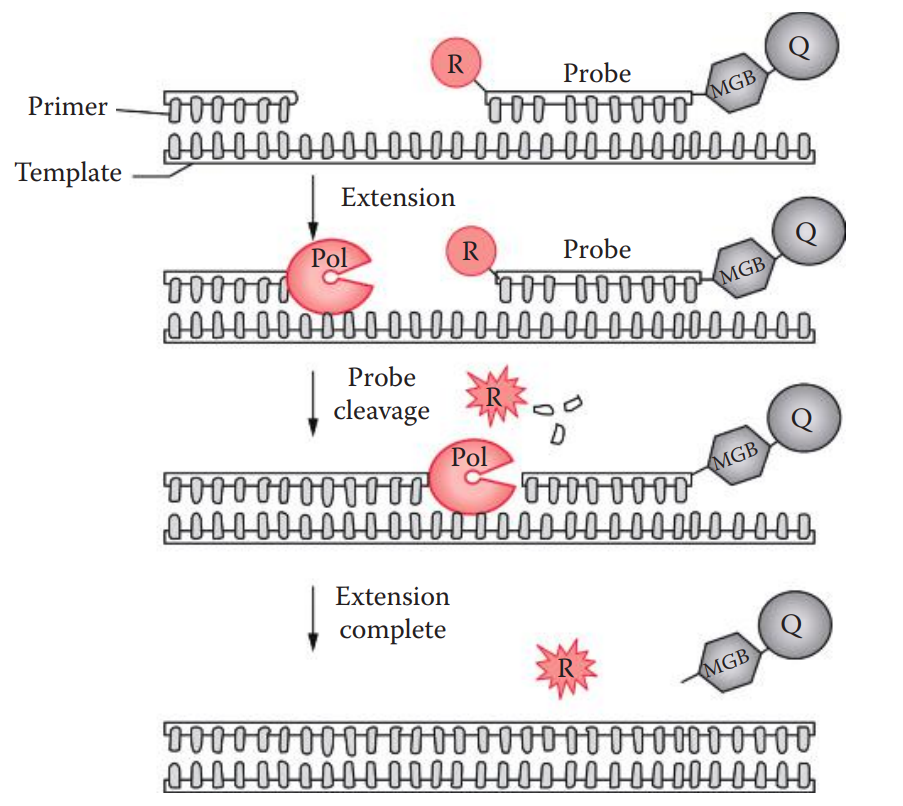
TaqMan Method
This method utilizes the 5′ exonuclease activity of Taq polymerase to cleave the probe during PCR.
A minor groove binder (MGB) is often linked at the 3′ ends of the probe.
A conjugated MGB binds to the minor groove of a B-form DNA helix which is stabilized by Vander Waals forces.
The oligonucleotide probe is labeled with both a reporter fluorescent dye, usually 6-carboxyfluorescein (6-FAM) or tetrachlorofluorescein (TET) at the 5′ ends; and a nonfluorescent quencher moiety, such as tetramethyl- rhodamine (TAMRA), usually at the 3′ ends or any thymine position.
Fluorescent Resonance Energy Transfers (FRET): A distance-dependent interaction between two molecules in which the excitation energy is transferred from a photon donor molecule (reporter) to an acceptor molecule (quencher) without emission of a photon.
Chapter 6: DNA Quantitation
6.1: Slot Blot Assay
Historically, the slot blot assay was used to detect human genomic DNA in a sample.
Prior to the quantitation, an alkaline solution is added to the genomic DNA sample, which denatures DNA.
Generating single-stranded DNA is necessary for DNA to be cross-linked onto a nitrocellulose membrane.
Three detecting schemes of the slot blot assay have been developed:
D17Z1 probe: Labeled with radioisotopes that could be visualized by exposing the slot blot membrane to x-ray film.
Alkaline phosphatase-labeled probe: It can be coupled with chemiluminescent detection.
Biotinylated probe: It can be coupled with either colorimetric or chemiluminescent detection.
In colorimetric detection, the biotin moiety of the probe is bound to streptavidin.
The streptavidin is conjugated with horseradish peroxidase, which catalyzes the oxidation reaction of tetramethylbenzidine (TMB), a substrate, forming a blue precipitate.
With the chemiluminescent detection method, the horseradish peroxidase catalyzes the oxidation reaction of a substrate such as luminol, emitting photons that can be detected by exposure to x-ray film.
The sensitivity of chemiluminescent detection is slightly higher than that of colorimetric detection.
The detected signal intensity is proportional to the concentration of the DNA sample in question. Quantitative measurements can be made by comparing an unknown sample to a set of standards with known DNA concentrations.
6.2: Fluorescent Intercalating Dye Assay
Intercalating dyes, usually planar molecules, can slide themselves in between base pairs of DNA without breaking the DNA double helix.
The Quant-iT PicoGreen dsDNA reagent (Invitrogen) is a fluorescent intercalating dye that stains double-stranded DNA (dsDNA) for quantitation in a sample. The detection limit of this method is approximately 250 pg.
Fluorescent intercalating dye assay can be utilized for the quantitation of known reference samples.
DNA samples are simply added to a solution containing the fluorescent intercalating dye. The fluorescence, proportional to the quantities of DNA is measured using a standard spectrofluorometer with excitation and emission wavelengths of the light source.
A standard curve is first created using samples containing known amounts of DNA. The assay is then performed for the unknown samples and the quantities of DNA in the samples are determined by comparing the results to the standard curve.




6.3: Quantitative PCR Assay
End-point PCR methods measure the quantity of amplified product at the end of PCR.
Real-time PCR methods can quantify the amplified DNA during the exponential phase of PCR.
Real-Time Quantitative PCR
Real-time quantitative PCR (qPCR) was developed in the early 1990s, and it analyzes the amplification of a target sequence at each cycle of PCR.
Fluorescent reporter: Used to monitor the accumulation of amplified products during PCR.
The fluorescence signals of the reporter molecule increase as amplified products accumulate with each cycle of PCR.
qPCR is commonly used because of the following advantages:
Better objectivity than the QuantiBlot method.
Increased sensitivity with a large dynamic range (30 pg–100 ng).
More accurate measurements of small quantities of DNA in samples.
Fewer laboratory manipulations; are amenable to automation.
Ability to detect PCR inhibitors.
The technique is amenable to multiplexing to detect more than one type of DNA target sequence in a single reaction.

TaqMan Method
This method utilizes the 5′ exonuclease activity of Taq polymerase to cleave the probe during PCR.
A minor groove binder (MGB) is often linked at the 3′ ends of the probe.
A conjugated MGB binds to the minor groove of a B-form DNA helix which is stabilized by Vander Waals forces.
The oligonucleotide probe is labeled with both a reporter fluorescent dye, usually 6-carboxyfluorescein (6-FAM) or tetrachlorofluorescein (TET) at the 5′ ends; and a nonfluorescent quencher moiety, such as tetramethyl- rhodamine (TAMRA), usually at the 3′ ends or any thymine position.
Fluorescent Resonance Energy Transfers (FRET): A distance-dependent interaction between two molecules in which the excitation energy is transferred from a photon donor molecule (reporter) to an acceptor molecule (quencher) without emission of a photon.

 Knowt
Knowt