
Principles of Life, Chapter 12 Reading
12.1 The -omics Era has Revolutionized Biology
Forward Genetics: An approach that begins with an interesting phenotype, finds the gene(s) underlying it, and then determines as much as possible about those genes.
Reverse Genetics: An approach that begins with a gene and attempts to determine its function, often by examining what happens when the gene is knocked out.
Sanger dideoxy sequencing method
Statements made by the Stanger method
DNA polymerase requires a primer to begin polymerization
A 3’ OH group on the end nucleotide must be available on the growing strand for the addition of the next nucleotide
A single-stranded DNA primer is designed to be the starting point for DNA synthesis on the strand to be sequenced
dNTPs are used
4 ddNTPs are added, each modified to floresce a certain color
Each lacks a 3’ OH so synthesis terminates when a ddNTP is incorporated
Product DNA varies in length, in one-nucleotide increments, labeled so the end nucleotide can be id’d
Sanger sequencing allows DNA sequences of 300 to 1k bases to be obtained in a single reaction
High-throughput sequencing: Rapid DNA sequencing on a micro scale in which many fragments of DNA are sequenced in parallel. Also known as next-generation sequencing.
About 500 million reactions can occur on a very small surface.
DNA is prepped for sequencing
Many copies of a large DNA molecule are cut into small 300bp each
Fragments are denatured by heat and short, synthetic adapter sequences (oligonucleotides) are then attached to each end of each fragment
Fragments are attachd to the surface of a solid support, leaving a small amount of space between each bolecule
Single stranded DNA primers complementary to the adapter sequenced are used in PCR
Templates are used for sequencing
DNA fragments are heated to denature them; universal primer, polymerase, and four dNTPs are added (NOT ddNTPs)
The sequencing reaction is set up so that only one nucleotide at a time is addd to the new DNA strand
Fluorescence of the newly added nucleotide at each location is detected with a camera (color indicates which nucleotide was added)
Repeated with only more florescent nucleotide added during each replication; images are taken after each one.
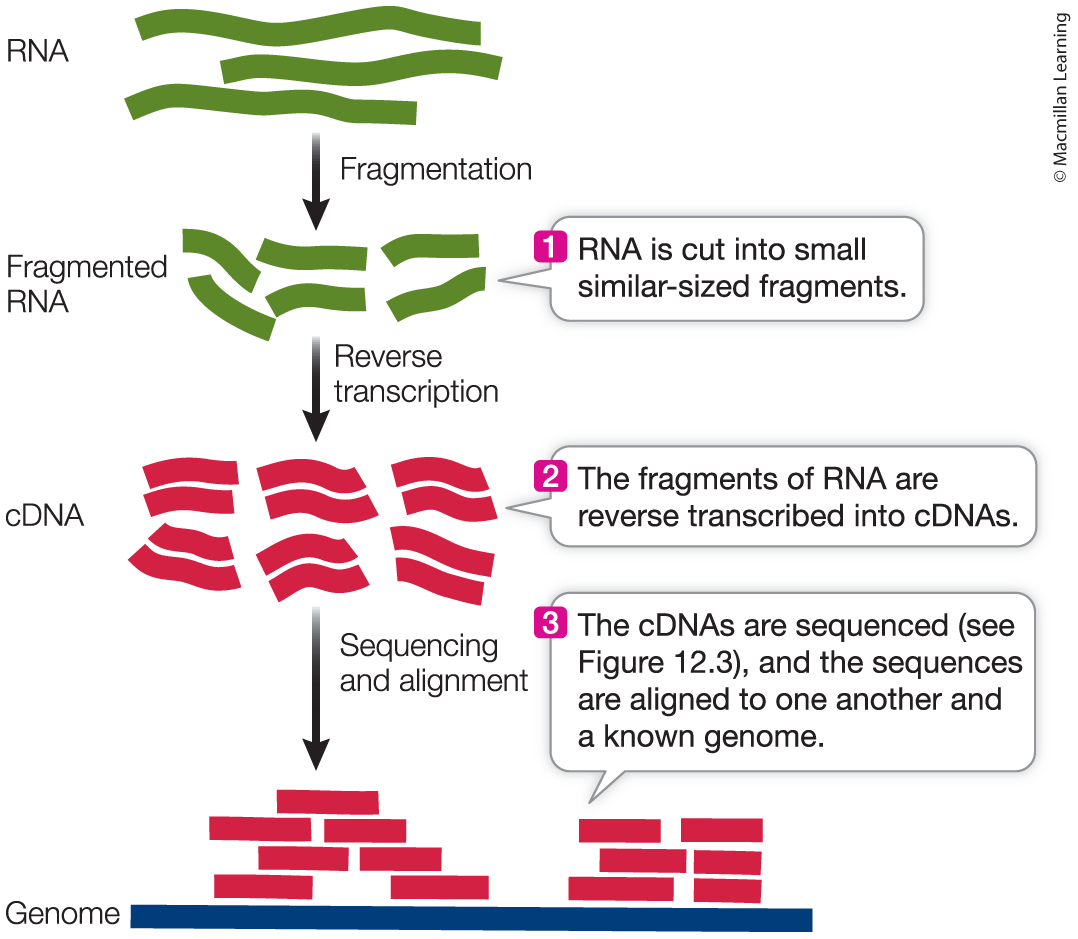
Transcriptomics identifies expressed genes
A genome is much larger than the transcriptome
Transcriptome: The subset of the genome that is expressed as RNA in a particular cell or tissue at a particular time.
Better because only the expressed genes are likely to affect phenotype
The genomes of each somatic cell are identical, but their transcriptomes are not
RNA Sequencing: The sequencing of all of the complementary DNA in a sample, obtained from the RNA by reverse transcription.
Genome sequences yield several kinds of information
Open reading frames: Sequences of DNA within genes that begin with a start codon and end with a stop codon.
An open reading frame that begins with an intron consensus sequence (boundary between exon and intron) and ends with an intron consensus sequence may be an internal exon.
After identifying the proteins
Promteomics: The study of the proteome—the complete complement of proteins produced by an organism.
Separation based on size/pH
Mass spectrometry
Antibodies
Metabolomics: The study of the metabolome (complete set of small molecules present) as it relates to the physiological state of a cell, tissue, or organism.
Primary: normal cellular process molecules
Secondary: unique to specific groups of organisms

12.2 Prokaryotic Genomes are Small, Compact, and Diverse
Bacterial genomes are small, compact, usually have no introns, and carry plasmids along with their main chromosome
The number of genes shared across all prokaryotes is quite small (implies you only need a few for life)
Core genome: The part of a genome found in all individuals (or strains) within a species.
Pan genome: The entirety of genome sequence found across all individuals (or strains) within a species.
Metagenomics: The practice of analyzing DNA from environmental samples without isolating intact organisms.
Transposons: Mobile DNA segments that can insert into a chromosome and cause genetic change; do not use RNA intermediates; pretty much junk DNA

12.3 Eukaryotic Genomes are Large and Complex
Eukaryotic genomes are much larger, have more regulatory sequences, and most DNA does not encode functional proteins, and the core/pan genomes are usually the same.
Yeast
Single-celled eukaryotic microbe and is the least complex of the eukaryotic model organisms.
Lives like a prokaryote but has membrane-enclosed organelles like other eukaryotes.
Only 16 linear chromosomes and 6,600 protein-coding genes.
Only 20 percent or so are required for growth on rich medium.
It is the compartmentalization of the eukaryotic yeast cell into organelles that requires it to have more genes.
Confirms that the eukaryotic cell is structurally and functionally more complex than the prokaryotic cell.
Nematode
Only 1,000 or so somatic cells in an adult and a transparent body.
Develops over 3 days from a fertilized egg to an adult worm that has a nervous system, digests food, and reproduces sexually.
8 times larger than that of yeast and has about three times as many protein-coding genes.
Many of these extra genes encode proteins needed for cell differentiation, for intercellular communication, and for holding cells together to form tissues.
Most of the genes in the nematode are not essential—only 30%
Orthologs: Homologous genes whose divergence can be traced to speciation events.
Pseudogene: A DNA segment that is homologous to a functional gene but is not expressed because of changes to its sequence or changes to its location in the genome.
Paralogs: Homologous genes whose divergence can be traced to gene duplication events.
Gene family: A set of similar genes derived from a single parent gene; need not be on the same chromosomes. The vertebrate globin genes constitute a classic example of a gene family.
Human Genome
1% or 2% percent of the genome encoding protein-coding sequences
39% encodes genes or gene-related sequences. These include introns (30 percent of the genome) and regulatory regions, including promoters, genes, and pseudogenes.
60% of the human genome that is intergenic (not gene-related).
Miscrosatellites: Simple 1–5 base pair DNA repeats, present in multiple tandem copies; also known as STRs or SSRs
Retrotransposons: Mobile genetic elements that are reverse transcribed into RNA as part of their transfer mechanism.
Long Terminal Repeats Retrotransposons have these LTRs on both ends of the DNA; are 8% of human genome
Non LTR Retrortansposons have no LTR at their ends
SINEs (short interspersed elements) are transcribed but not translated (13% of total genome)
LINEs (ling interspered elements) some are transcribed and translated; 20% of human genome
12.3 The Human Genome
Among the 3.1 billion bp in the haploid human genome, there are about 20,500 protein-coding genes.
Means that posttranscriptional mechanisms (such as alternative splicing) must account for the observed number of proteins in humans. It turns out that most human genes encode multiple proteins via alternative splicing.
There are another 24,000 non-protein-coding genes that are expressed in RNA
Suggests their importance in processes such as posttranscriptional gene regulation
The median size of a protein-coding gene is about 26,000 bp, and virtually all genes have many introns (the median number is 7).
Approximately half of the genome is made up of transposons and other repetitive sequences
Most transposons are inactive most of the time.
Most of the genome (at least 99 percent) is the same in all people.
Within an individual, the genomes of different somatic cell lines will diverge as they get new mutations.

Single nucleotide polymorphisms (SNPs): Inherited variations in a single nucleotide base in DNA that differ between individuals.
Used for ancestry
Determine genetic causes of quantitative traits
DNA Microarray: A small glass or plastic square onto which thousands of single-stranded DNA sequences are fixed so that hybridization of cell-derived RNA or DNA to the target sequences can be performed.
Nucleic Acid Hybridization: A technique in which a single-stranded nucleic acid probe is made that is complementary to, and binds to, a target sequence, either DNA or RNA. The resulting double-stranded molecule is a hybrid.
Personal genomics: The use of an individual’s genome sequence to inform ancestry determination, risks of genetic disease and response to drugs.
Pharmacogenomics: The study of how an individual’s genetic makeup affects their response to drugs or other agents, with the goal of predicting the effectiveness of different treatment options.
Principles of Life, Chapter 12 Reading
12.1 The -omics Era has Revolutionized Biology
Forward Genetics: An approach that begins with an interesting phenotype, finds the gene(s) underlying it, and then determines as much as possible about those genes.
Reverse Genetics: An approach that begins with a gene and attempts to determine its function, often by examining what happens when the gene is knocked out.
Sanger dideoxy sequencing method
Statements made by the Stanger method
DNA polymerase requires a primer to begin polymerization
A 3’ OH group on the end nucleotide must be available on the growing strand for the addition of the next nucleotide
A single-stranded DNA primer is designed to be the starting point for DNA synthesis on the strand to be sequenced
dNTPs are used
4 ddNTPs are added, each modified to floresce a certain color
Each lacks a 3’ OH so synthesis terminates when a ddNTP is incorporated
Product DNA varies in length, in one-nucleotide increments, labeled so the end nucleotide can be id’d
Sanger sequencing allows DNA sequences of 300 to 1k bases to be obtained in a single reaction
High-throughput sequencing: Rapid DNA sequencing on a micro scale in which many fragments of DNA are sequenced in parallel. Also known as next-generation sequencing.
About 500 million reactions can occur on a very small surface.
DNA is prepped for sequencing
Many copies of a large DNA molecule are cut into small 300bp each
Fragments are denatured by heat and short, synthetic adapter sequences (oligonucleotides) are then attached to each end of each fragment
Fragments are attachd to the surface of a solid support, leaving a small amount of space between each bolecule
Single stranded DNA primers complementary to the adapter sequenced are used in PCR
Templates are used for sequencing
DNA fragments are heated to denature them; universal primer, polymerase, and four dNTPs are added (NOT ddNTPs)
The sequencing reaction is set up so that only one nucleotide at a time is addd to the new DNA strand
Fluorescence of the newly added nucleotide at each location is detected with a camera (color indicates which nucleotide was added)
Repeated with only more florescent nucleotide added during each replication; images are taken after each one.
Transcriptomics identifies expressed genes
A genome is much larger than the transcriptome
Transcriptome: The subset of the genome that is expressed as RNA in a particular cell or tissue at a particular time.
Better because only the expressed genes are likely to affect phenotype
The genomes of each somatic cell are identical, but their transcriptomes are not
RNA Sequencing: The sequencing of all of the complementary DNA in a sample, obtained from the RNA by reverse transcription.

Genome sequences yield several kinds of information
Open reading frames: Sequences of DNA within genes that begin with a start codon and end with a stop codon.
An open reading frame that begins with an intron consensus sequence (boundary between exon and intron) and ends with an intron consensus sequence may be an internal exon.
After identifying the proteins
Promteomics: The study of the proteome—the complete complement of proteins produced by an organism.
Separation based on size/pH
Mass spectrometry
Antibodies
Metabolomics: The study of the metabolome (complete set of small molecules present) as it relates to the physiological state of a cell, tissue, or organism.
Primary: normal cellular process molecules
Secondary: unique to specific groups of organisms

12.2 Prokaryotic Genomes are Small, Compact, and Diverse
Bacterial genomes are small, compact, usually have no introns, and carry plasmids along with their main chromosome
The number of genes shared across all prokaryotes is quite small (implies you only need a few for life)
Core genome: The part of a genome found in all individuals (or strains) within a species.
Pan genome: The entirety of genome sequence found across all individuals (or strains) within a species.
Metagenomics: The practice of analyzing DNA from environmental samples without isolating intact organisms.
Transposons: Mobile DNA segments that can insert into a chromosome and cause genetic change; do not use RNA intermediates; pretty much junk DNA

12.3 Eukaryotic Genomes are Large and Complex
Eukaryotic genomes are much larger, have more regulatory sequences, and most DNA does not encode functional proteins, and the core/pan genomes are usually the same.
Yeast
Single-celled eukaryotic microbe and is the least complex of the eukaryotic model organisms.
Lives like a prokaryote but has membrane-enclosed organelles like other eukaryotes.
Only 16 linear chromosomes and 6,600 protein-coding genes.
Only 20 percent or so are required for growth on rich medium.
It is the compartmentalization of the eukaryotic yeast cell into organelles that requires it to have more genes.
Confirms that the eukaryotic cell is structurally and functionally more complex than the prokaryotic cell.
Nematode
Only 1,000 or so somatic cells in an adult and a transparent body.
Develops over 3 days from a fertilized egg to an adult worm that has a nervous system, digests food, and reproduces sexually.
8 times larger than that of yeast and has about three times as many protein-coding genes.
Many of these extra genes encode proteins needed for cell differentiation, for intercellular communication, and for holding cells together to form tissues.
Most of the genes in the nematode are not essential—only 30%
Orthologs: Homologous genes whose divergence can be traced to speciation events.
Pseudogene: A DNA segment that is homologous to a functional gene but is not expressed because of changes to its sequence or changes to its location in the genome.
Paralogs: Homologous genes whose divergence can be traced to gene duplication events.
Gene family: A set of similar genes derived from a single parent gene; need not be on the same chromosomes. The vertebrate globin genes constitute a classic example of a gene family.
Human Genome
1% or 2% percent of the genome encoding protein-coding sequences
39% encodes genes or gene-related sequences. These include introns (30 percent of the genome) and regulatory regions, including promoters, genes, and pseudogenes.
60% of the human genome that is intergenic (not gene-related).
Miscrosatellites: Simple 1–5 base pair DNA repeats, present in multiple tandem copies; also known as STRs or SSRs
Retrotransposons: Mobile genetic elements that are reverse transcribed into RNA as part of their transfer mechanism.
Long Terminal Repeats Retrotransposons have these LTRs on both ends of the DNA; are 8% of human genome
Non LTR Retrortansposons have no LTR at their ends
SINEs (short interspersed elements) are transcribed but not translated (13% of total genome)
LINEs (ling interspered elements) some are transcribed and translated; 20% of human genome
12.3 The Human Genome
Among the 3.1 billion bp in the haploid human genome, there are about 20,500 protein-coding genes.
Means that posttranscriptional mechanisms (such as alternative splicing) must account for the observed number of proteins in humans. It turns out that most human genes encode multiple proteins via alternative splicing.
There are another 24,000 non-protein-coding genes that are expressed in RNA
Suggests their importance in processes such as posttranscriptional gene regulation
The median size of a protein-coding gene is about 26,000 bp, and virtually all genes have many introns (the median number is 7).
Approximately half of the genome is made up of transposons and other repetitive sequences
Most transposons are inactive most of the time.
Most of the genome (at least 99 percent) is the same in all people.
Within an individual, the genomes of different somatic cell lines will diverge as they get new mutations.

Single nucleotide polymorphisms (SNPs): Inherited variations in a single nucleotide base in DNA that differ between individuals.
Used for ancestry
Determine genetic causes of quantitative traits
DNA Microarray: A small glass or plastic square onto which thousands of single-stranded DNA sequences are fixed so that hybridization of cell-derived RNA or DNA to the target sequences can be performed.
Nucleic Acid Hybridization: A technique in which a single-stranded nucleic acid probe is made that is complementary to, and binds to, a target sequence, either DNA or RNA. The resulting double-stranded molecule is a hybrid.
Personal genomics: The use of an individual’s genome sequence to inform ancestry determination, risks of genetic disease and response to drugs.
Pharmacogenomics: The study of how an individual’s genetic makeup affects their response to drugs or other agents, with the goal of predicting the effectiveness of different treatment options.
 Knowt
Knowt
