
Chapter 23: Mitochondrial DNA Profiling
23.1: Human Mitochondrial Genome
Mitochondria: Are cellular organelles that serve as the energy-generating components of cells.
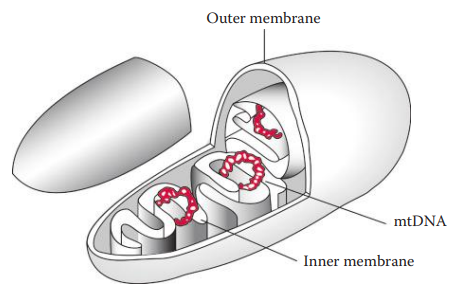
Genetic Contents of Mitochondrial Organelle Genomes
Cambridge reference sequence (CRS): The sequence was largely derived from a placental sample from an individual of European descent and also partially from HeLa cells and bovine cells.
It was later discovered, by resequencing the original mtDNA sample, that the CRS contains substitution errors at 10 nucleotide positions.
The revised Cambridge reference sequence (rCRS) was published in 1999 and presented corrections to these substitution errors.
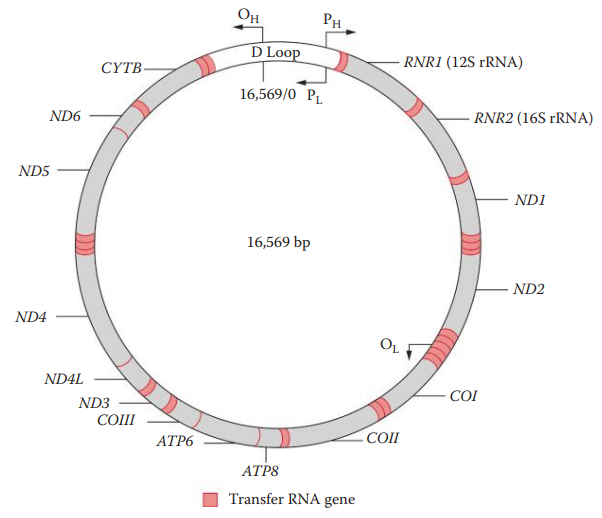
Control Region: also known as Displacement Loop; it contains the origin of replication for one of the mtDNA strands but does not code for any gene products.
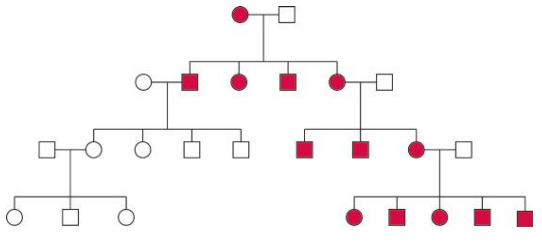
Maternal Inheritance of mtDNA
Maternal inheritance is typically observed for the mtDNA genome, which is inherited differently from nuclear genes.
The inheritance of the mtDNA genome does not obey the rules of Mendelian inheritance and is thus called non-Mendelian inheritance.
Mitotype: Considered a a haplotype treated as a single locus.
23.2: mtDNA Polymorphic Regions
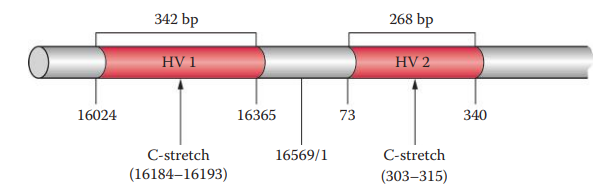
Hypervariable Regions
The most polymorphic region of mtDNA is located within the D-loop.
The three hypervariable regions in the D-loop are designated: Hypervariable Region I, Hypervariable Region II and, Hypervariable Region III
The most common polymorphic regions of the human mtDNA genome analyzed for forensic purposes are HV1 and HV2.
Heteroplasmy
Heteroplasmy occurs when an individual carries more than one mtDNA haplotype.
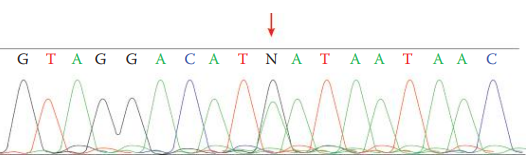
Sequence heteroplasmy: The presence of two different nucleotides at a single position shown as overlapping peaks in a sequence electropherogram.
Light heteroplasmy: Are often observed at the uninterrupted C stretches in sequencing, in which sequencing products with various lengths of polymeric cytosine residues are present.
23.3: Forensic mtDNA Testing
General Considerations
mtDNA analysis is often used on samples derived from skeletal or decomposed remains.
The surface of the sample should be cleaned to remove any adhering debris or contaminants.
Bones and teeth are pulverized to facilitate extraction of the mtDNA Assay.
mtDNA-specific quantization methods using real-time PCR can also be used to directly obtain measurements of mtDNA extracted.
mtDNA Screen Assay
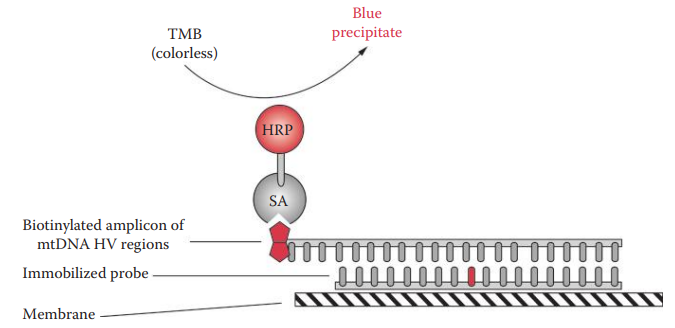
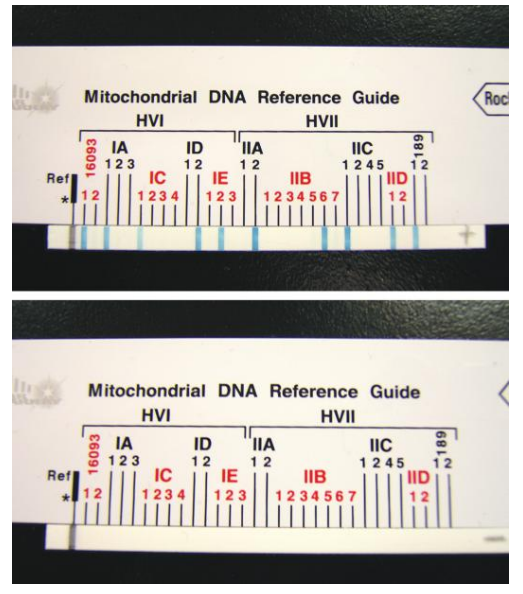
ASO Assay: It allows initial screening of mtDNA sequence polymorphisms and has the potential to reduce the number of samples required for mtDNA sequencing.
Linear Array™ mtDNA HV1/HV2 region sequence typing kit: It utilizes reverse ASO configuration with a panel of immobilized ASO probes that detect common polymorphic sites.
mtDNA Sequencing
A combination of PCR amplification and DNA sequencing techniques is utilized to reduce the time and labor needed to obtain DNA sequences from genomic DNA templates.
mtDNA sequencing usually consists of:
PCR amplification;
DNA sequencing reactions;
Separation using electrophoresis; and
Data collection of Sequence analysis.
DNA Sequencing Reactions
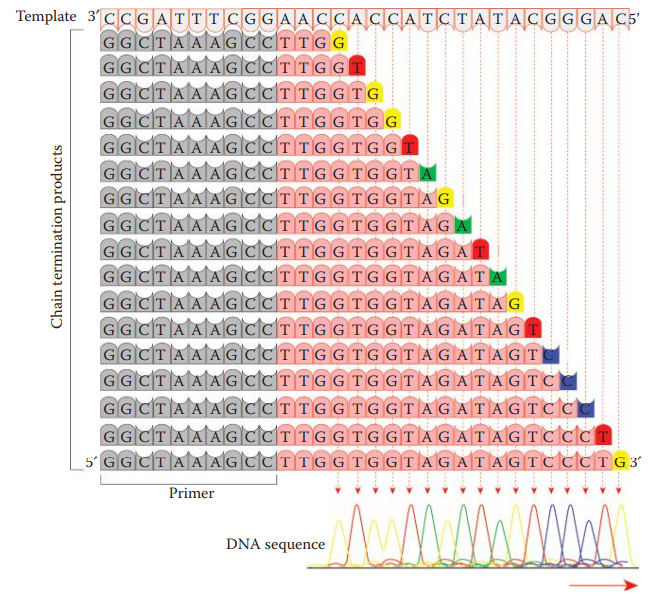
Chain-Termination or Sanger Method: An oligonucleotide primer that can anneal to a single stranded DNA template is utilized.
Cycle Sequencing: It utilizes thermal cycling to generate a single-stranded template for chain-termination sequencing reactions.
Electrophoresis, Sequence Analysis, and Mitotype Designations
Reporting Format: Sequence differences relative to the rCRS are listed in data format.
Insertions: Described by noting the position followed by a decimal point and a number.
Deletions: These site designation is followed by letter d.
Hetero-plasmic Sites: The IUPAC codes for base calling can be applied to hetero-plasmic sites.
Interpretation of mtDNA Profiling Results
Interpretation guidelines are used when comparing sequencing results between evidence and reference samples.
Exclusion: If the sequences of questioned and known samples are different, then the samples can be excluded as originating from the same source.
It should be taken into account that higher mutation rates are found with the mtDNA genome than are found with the nuclear genome.
Cannot Exclude: If the sequences are the same, the reference sample and evidence cannot be excluded as arising from the same source.
When an mtDNA profile cannot be excluded, it is desirable to evaluate the weight of the evidence.
Inconclusive Result: If the questioned and known samples differ by a single nucleotide, and no heteroplasmy is present, the results are considered to be inconclusive.
Chapter 23: Mitochondrial DNA Profiling
23.1: Human Mitochondrial Genome
Mitochondria: Are cellular organelles that serve as the energy-generating components of cells.

Genetic Contents of Mitochondrial Organelle Genomes
Cambridge reference sequence (CRS): The sequence was largely derived from a placental sample from an individual of European descent and also partially from HeLa cells and bovine cells.
It was later discovered, by resequencing the original mtDNA sample, that the CRS contains substitution errors at 10 nucleotide positions.
The revised Cambridge reference sequence (rCRS) was published in 1999 and presented corrections to these substitution errors.
Control Region: also known as Displacement Loop; it contains the origin of replication for one of the mtDNA strands but does not code for any gene products.


Maternal Inheritance of mtDNA
Maternal inheritance is typically observed for the mtDNA genome, which is inherited differently from nuclear genes.
The inheritance of the mtDNA genome does not obey the rules of Mendelian inheritance and is thus called non-Mendelian inheritance.
Mitotype: Considered a a haplotype treated as a single locus.
23.2: mtDNA Polymorphic Regions
Hypervariable Regions
The most polymorphic region of mtDNA is located within the D-loop.
The three hypervariable regions in the D-loop are designated: Hypervariable Region I, Hypervariable Region II and, Hypervariable Region III
The most common polymorphic regions of the human mtDNA genome analyzed for forensic purposes are HV1 and HV2.


Heteroplasmy
Heteroplasmy occurs when an individual carries more than one mtDNA haplotype.
Sequence heteroplasmy: The presence of two different nucleotides at a single position shown as overlapping peaks in a sequence electropherogram.
Light heteroplasmy: Are often observed at the uninterrupted C stretches in sequencing, in which sequencing products with various lengths of polymeric cytosine residues are present.

23.3: Forensic mtDNA Testing
General Considerations
mtDNA analysis is often used on samples derived from skeletal or decomposed remains.
The surface of the sample should be cleaned to remove any adhering debris or contaminants.
Bones and teeth are pulverized to facilitate extraction of the mtDNA Assay.
mtDNA-specific quantization methods using real-time PCR can also be used to directly obtain measurements of mtDNA extracted.
mtDNA Screen Assay
ASO Assay: It allows initial screening of mtDNA sequence polymorphisms and has the potential to reduce the number of samples required for mtDNA sequencing.
Linear Array™ mtDNA HV1/HV2 region sequence typing kit: It utilizes reverse ASO configuration with a panel of immobilized ASO probes that detect common polymorphic sites.
mtDNA Sequencing
A combination of PCR amplification and DNA sequencing techniques is utilized to reduce the time and labor needed to obtain DNA sequences from genomic DNA templates.
mtDNA sequencing usually consists of:
PCR amplification;
DNA sequencing reactions;
Separation using electrophoresis; and
Data collection of Sequence analysis.
DNA Sequencing Reactions
Chain-Termination or Sanger Method: An oligonucleotide primer that can anneal to a single stranded DNA template is utilized.
Cycle Sequencing: It utilizes thermal cycling to generate a single-stranded template for chain-termination sequencing reactions.
Electrophoresis, Sequence Analysis, and Mitotype Designations
Reporting Format: Sequence differences relative to the rCRS are listed in data format.
Insertions: Described by noting the position followed by a decimal point and a number.
Deletions: These site designation is followed by letter d.
Hetero-plasmic Sites: The IUPAC codes for base calling can be applied to hetero-plasmic sites.



Interpretation of mtDNA Profiling Results
Interpretation guidelines are used when comparing sequencing results between evidence and reference samples.
Exclusion: If the sequences of questioned and known samples are different, then the samples can be excluded as originating from the same source.
It should be taken into account that higher mutation rates are found with the mtDNA genome than are found with the nuclear genome.
Cannot Exclude: If the sequences are the same, the reference sample and evidence cannot be excluded as arising from the same source.
When an mtDNA profile cannot be excluded, it is desirable to evaluate the weight of the evidence.
Inconclusive Result: If the questioned and known samples differ by a single nucleotide, and no heteroplasmy is present, the results are considered to be inconclusive.
 Knowt
Knowt
